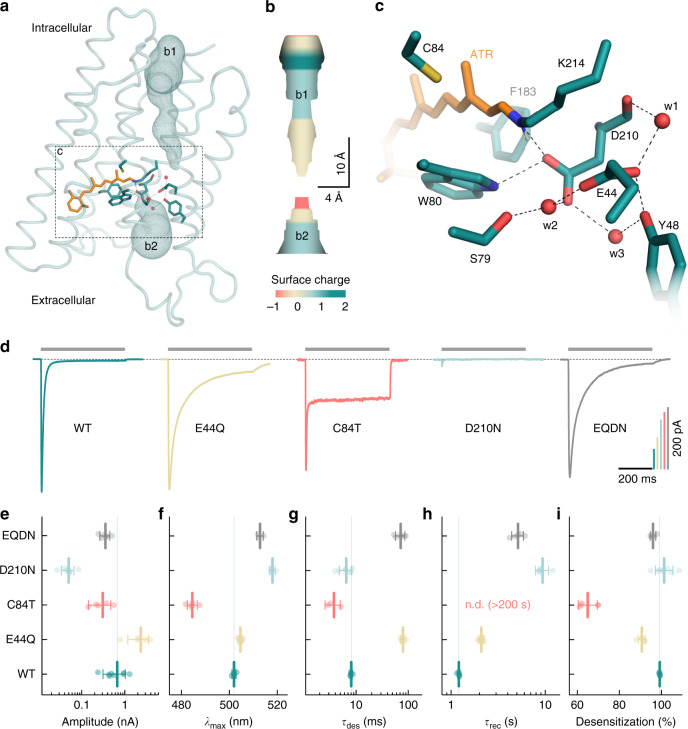
Fig. 4.
MD simulations and mutational analysis of MerMAID1. a Overview of the MD simulation homology model of MerMAID1 in the dark. The predicted ion permeation pathway is shown as mesh (b1, b2), and ribbons represent the protein backbone. b Electrostatic surface potential of the predicted chloride permeation pathway. c Detailed view of the active-site residues, with amino acids shown as cyan sticks and the all-trans retinal (ATR) in orange. Red spheres denote water molecules that remained stable during MD simulation. d Representative photocurrent traces of wild-type (WT) MerMAID1 and selected MerMAID1 mutants recorded at −60 mV. Photocurrent amplitudes (e), λmax (f), apparent τdes of the peak current (g), recovery time constant, τrec (h), and extent of desensitization (i) of WT MerMAID1 and indicated mutants. Mean values (thick lines) ± standard deviation (whiskers) are shown, and single-measurement data points are represented as dots. Source data are provided as a Source Data file (e–i)