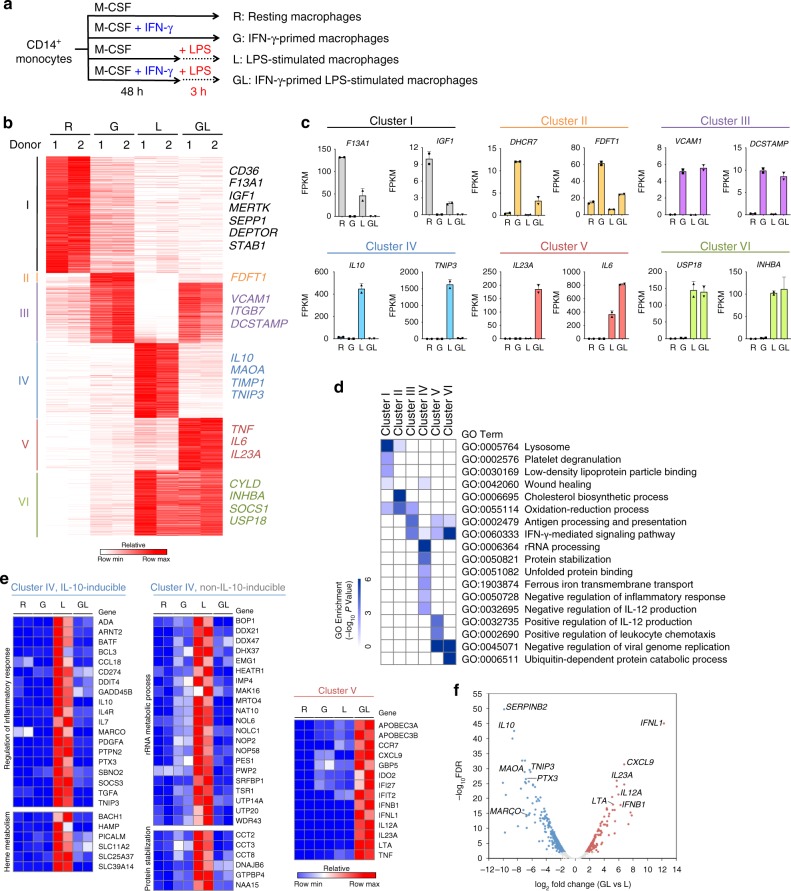
Fig. 1.
IFN-γ-mediated reprogramming of TLR4-induced transcriptome in human macrophages. a Experimental design: human CD14+ monocyte-derived macrophages underwent one of four different combinations of priming for 48 h with IFN-γ before stimulation with LPS (or not) for 3 h: without priming or stimulation (R); primed with IFN-γ without LPS stimulation (G); LPS stimulation only with no priming (L); or primed with IFN-γ and stimulated with LPS (GL). b K-means (K = 6) clustering of 3,909 differentially expressed genes in any pairwise comparison among four conditions. Clusters are indicated on the left. c Examples of expression of select genes from the six clusters identified in a. Each dot in the bar plot represents one donor and error bars represent standard deviation. d Heatmap showing the P-value significance of GO term enrichment for genes in each cluster. e Heatmap showing the relative expression of representative cluster IV genes that are inducible by IL-10 (left), cluster IV genes that are not inducible by IL-10 (middle), and cluster V genes. Distinct biological functions are indicated on the left. IL-10-inducible genes were obtained from GSE43700. f Volcano plot of transcriptomic changes between LPS (L) and IFN-γ-primed LPS-stimulated (GL) macrophages; colored dots correspond to genes with significant (FDR < 0.01) and greater than two-fold expression changes. Data from two independent experiments with different donors are depicted