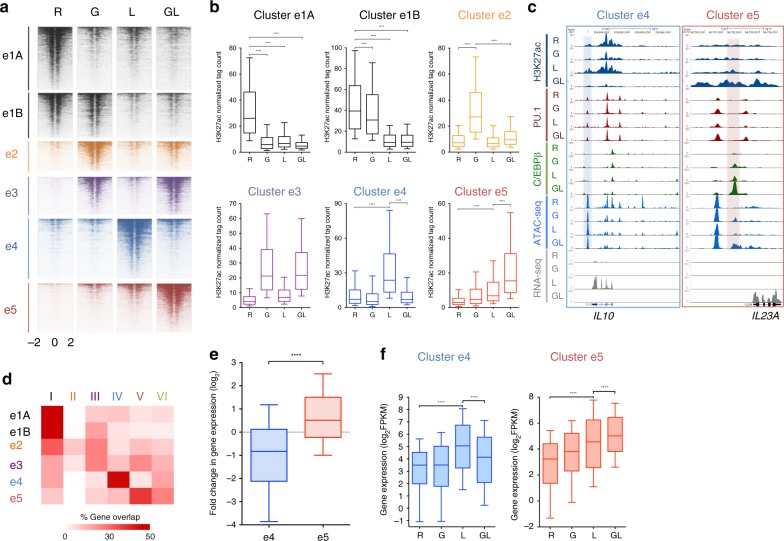
Fig. 2.
Selective regulation of LPS-activated enhancer landscapes by IFN-γ. a K-means clustering (K = 5) of enhancers as shown in Supplementary Fig. 2a further filtered (FDR < 0.05, > 2-fold changes) and subdivided into six enhancer clusters. Heatmaps showing H3K27ac ChIP-seq signals at each enhancer cluster. b The boxplots indicate normalized tag counts at each enhancer cluster. ****p < 0.0001, paired-samples Wilcoxon signed-rank test. Boxes encompass the twenty-fifth to seventy-fifth percentile changes. Whiskers extend to the tenth and nintieth percentiles. The central horizontal bar indicates the median. Data are representative of two independent experiments. c Representative UCSC Genome Browser tracks displaying normalized tag-density profiles at enhancers of IL10 and IL23A in four conditions. Shaded boxes enclose a cluster e4 enhancer (left) and cluster e5 enhancer (right). d Heatmap presentation of the percentage of genes in each cluster (Fig. 1a) that overlap with genes associated with the differentially regulated enhancer clusters identified in panel a. e Boxplots showing the change in gene expression between LPS and IFN-γ-primed and LPS-stimulated macrophages for the differentially expressed genes nearest (within 100 kb) to cluster e4 or e5 enhancers. ****p < 0.0001 by Welch’s t-test. f Boxplots showing gene expression in the four indicated conditions for e4- or e5-associated genes. ****p < 0.0001 by paired-samples Wilcoxon signed-rank test. Boxes encompass the twenty-fifth to seventy-fifth percentile changes. Whiskers extend to the tenth and nintieth percentiles. The central horizontal bar indicates the median. Data are representative of two independent experiments