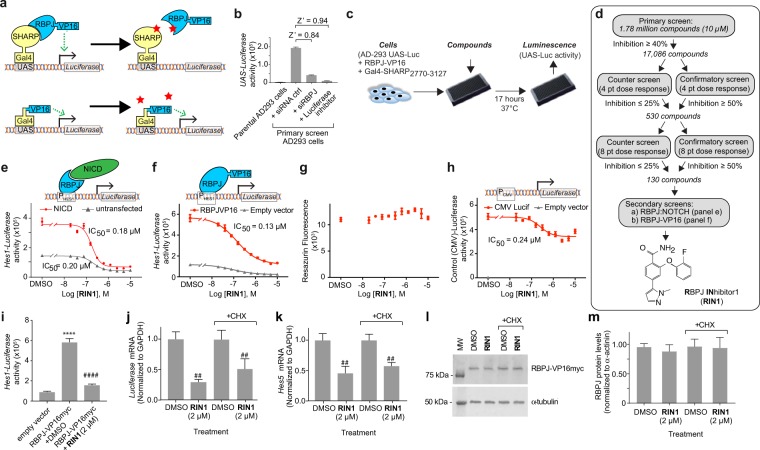
Figure 1.
Identification of small molecule RBPJ inhibitor, RIN1. (a) Schematic of the primary and counter screens. The screen was a cell-based two hybrid assay in which an active compound (stars) would disrupt the SHARP:RBPJ interaction and decrease activity of the Luciferase reporter. A minimal RBPJ-interacting domain of SHARP and a DNA-binding mutant of RBPJ were used (see Methods). (b) Assay validation using RBPJ siRNA transfection and a small molecule Luciferase inhibitor, Data is mean ± standard deviation, n = 5 wells. Z’ is a metric of dynamic range19. (c) Workflow schematic. (d) Screen flowchart and structure of RIN1. (e,f) Inhibition of NOTCH2 ICD (e) and RBPJ-VP16myc fusion protein (f) activity on the Hes1-Luciferase reporter in transient transfections, n = 4 wells. (g) Effect on cell viability, n = 4 wells. (h) Effect on CMV promoter activity, n = 4 wells. Data in b-h are presented as mean ± standard deviation; experiments were repeated > 3 times. (i–k) Effect of Cycloheximide on RIN1 inhibition of RBPJ-VP16. AD-293 cells were transfected with RBPJ-VP16myc and 48 hours later were treated ± RIN1 (2 µM), ± Cycloheximide (CHX, 10 µg/ml) for an additional 17 hours and then assayed for Hes1-Luciferase activity (i) (n = 10 wells), Luciferase mRNA (j) and endogenous HES5 mRNA (k), (n = 3 samples). Western blot of RBPJ-VP16myc fusion protein under identical conditions (l) and its quantification (m), n = 3 wells. Data are presented as mean ± standard deviation. Incrementing number of symbols (* and #) denote P < 0.05, P < 0.01, P < 0.001 and P < 0.0001 respectively, using two-tailed unpaired Student’s T-test relative to empty vector (*) or to RBPJ-VP16myc + DMSO vehicle control (#) conditions. Experiments were repeated twice.