Pythium insidiosum is an oomycete microorganism that causes a life-threatening infectious disease, called pythiosis, in humans and animals. The disease has been increasingly reported worldwide.
KEYWORDS: Pythium insidiosum, disulfiram, drug repurposing, pythiosis, susceptibility
ABSTRACT
Pythium insidiosum is an oomycete microorganism that causes a life-threatening infectious disease, called pythiosis, in humans and animals. The disease has been increasingly reported worldwide. Conventional antifungal drugs are ineffective against P. insidiosum. Treatment of pythiosis requires the extensive removal of infected tissue (i.e., eye and leg), but inadequate surgery and recurrent infection often occur. A more effective treatment is needed for pythiosis patients. Drug repurposing is a promising strategy for the identification of a U.S. Food and Drug Administration-approved drug for the control of P. insidiosum. Disulfiram has been approved to treat alcoholism, but it exhibits antimicrobial activity against various pathogens. In this study, we explored whether disulfiram possesses an anti-P. insidiosum activity. A total of 27 P. insidiosum strains, isolated from various hosts and geographic areas, were susceptible to disulfiram in a dose-dependent manner. The MIC range of disulfiram against P. insidiosum (8 to 32 mg/liter) was in line with that of other pathogens. Proteogenomic analysis indicated that several potential targets of disulfiram (i.e., aldehyde dehydrogenase and urease) were present in P. insidiosum. By homology modeling and molecular docking, disulfiram can bind the putative aldehyde dehydrogenase and urease of P. insidiosum at low energies (i.e., –6.1 and –4.0 Kcal/mol, respectively). Disulfiram diminished the biochemical activities of these enzymes. In conclusion, disulfiram can inhibit the growth of many pathogenic microorganisms, including P. insidiosum. The drug can bind and inactivate multiple proteins of P. insidiosum, which may contribute to its broad antimicrobial property. Drug repurposing of disulfiram could be a new treatment option for pythiosis.
INTRODUCTION
Pythium insidiosum is an oomycete microorganism that infects humans and animals and causes a life-threatening disease called pythiosis (1–3). Affected humans and animals have been increasingly reported worldwide, especially in the tropical and subtropical countries (3). Several serological and molecular assays have been developed to assist a rapid and accurate diagnosis of pythiosis (4–11). The use of conventional antifungal drugs is usually ineffective against P. insidiosum because the organism lacks the drug target enzymes in the sterol biosynthetic pathway (2, 12). Immunotherapy (i.e., administration of the P. insidiosum protein extract) shows limited efficacy in the treatment of pythiosis (2, 13). It requires the extensive removal of an infected organ (i.e., eye and leg) to eradicate the infection. However, adequate surgical intervention is not possible in many pythiosis patients, leading to recurrent infection and increased morbidity and mortality (2, 14). There is an urgent need for a more effective treatment for the control of P. insidiosum.
The development of a novel drug requires significant effort and time. In addition, diseases that are prevalent in lower-income countries receive less attention for drug development. Drug repurposing (or repositioning) is a strategy to identify an existing drug designed for one disease and use it in the treatment of a different disease (15–17). Success stories of the repurposing strategy include drug discoveries for infectious diseases. For example, auranofin (approved indication: rheumatoid arthritis), loperamide (diarrhea), tamoxifen (breast cancer), and chlorcyclizine (allergy) are repurposed as a new treatment option for infections caused by bacteria, parasites, fungi, or viruses (15). Drug repurposing is therefore a promising strategy for rapidly identifying a potential, ready-to-use, and safe compound (i.e., U.S. Food and Drug Administration [FDA]-approved drugs) for the control of other infectious diseases, including pythiosis.
Disulfiram is an FDA-approved drug used in the treatment of chronic alcoholism for decades (18). Several reports have shown that disulfiram markedly inhibits various pathogens, including Staphylococcus aureus (19), Pseudomonas aeruginosa (20), Giardia lamblia (21), Mycobacterium tuberculosis (22), and nontuberculous mycobacteria (23). The antimicrobial property of disulfiram could result from the inactivation of target enzymes, such as aldehyde dehydrogenase (ALDH), urease (URE), carbamate kinase (CK), and dopamine beta-hydroxylase (DBH) (21, 24–26). Because disulfiram possesses the broad-spectrum antimicrobial property, it is an open question whether the drug can be repurposed for the control of other pathogens, including P. insidiosum. In the present study, we performed an in vitro susceptibility assay and showed that all 27 P. insidiosum strains tested were susceptible to disulfiram. In addition, we identified some putative target enzymes in the genome of P. insidiosum (27, 28) and initially assessed a potential inhibitory mechanism of disulfiram against the pathogen by homology modeling, molecular docking, and biochemical analyses.
RESULTS
Susceptibility of P. insidiosum to disulfiram.
In this study, the P. insidiosum strains were isolated from various sources (i.e., humans, n = 15; animals, n = 9; and the environment, n = 3) and geographic origins (i.e., Asia, n = 20; Americas, n = 6; and Australia, n = 1) (Table 1). The ribosomal DNA (rDNA) sequence analysis assigned these strains into three genotypes: clade I (n = 5), clade II (n = 18), and clade III (n = 4) (29) (Table 1). The in vitro susceptibility assays (i.e., agar- and broth-based methods) showed that all 27 P. insidiosum strains were sensitive to disulfiram in a dose-dependent manner (Table 1; Fig. 1). The MICs of disulfiram against P. insidiosum varied, depending on the strains and susceptibility methods. The agar susceptibility method exhibited an MIC range of 16 to 128 mg/liter, an MIC50 of 32 mg/liter, and an MIC90 of 64 mg/liter. The broth susceptibility method showed an MIC range of 8 to 32 mg/liter, an MIC50 of 16 mg/liter, and an MIC90 of 32 mg/liter.
TABLE 1.
Twenty-seven strains of Pythium insidiosum used in the in vitro susceptibility assaysa
| Strain ID | Reference ID | Host (tissue) | Country of origin | Clade | Agar MIC (mg/liter) | Broth MIC (mg/liter) |
|---|---|---|---|---|---|---|
| Pi01 | CBS 578.85 | Equine | Costa Rica | I | 32 | 16 |
| Pi02 | CBS 579.85 | Equine | Costa Rica | I | 32 | 16 |
| Pi08 | CBS 580.85 | Equine | Costa Rica | I | 32 | 32 |
| Pi09 | CBS 101555 | Equine | Brazil | I | 32 | 32 |
| Pi10 | ATCC 200269 | Human (skin) | USA | I | 32 | 16 |
| Pi12 | SIMI 149-41 | Human (artery) | Thailand | II | 32 | 16 |
| Pi23 | MCC 10 | Human (gut) | Thailand | II | 32 | 32 |
| Pi25 | P19 | Human (eye) | Thailand | II | 32 | 16 |
| Pi29 | P27 | Human (artery) | Thailand | II | 32 | 16 |
| Pi36 | ATCC 64221 | Equine | Australia | II | 32 | 16 |
| Pi42 | CR02 | Environment | Thailand | II | 32 | 16 |
| Pi43 | LP04 | Environment | Thailand | II | 64 | 32 |
| Pi20 | MCC 18 | Human (eye) | Thailand | II | 32 | 8 |
| Pi35 | Pi-S | Human (artery) | Thailand | II | 128 | 8 |
| Pi33 | P36 | Human (artery) | Thailand | II | 32 | 32 |
| Pi34 | P37 | Human | Thailand | II | 32 | 32 |
| Pi52 | P38 | Human (artery) | Thailand | II | 64 | 32 |
| Pi53 | P39 | Equine (nose) | Thailand | II | 64 | 16 |
| Pi54 | P40 | Human (artery) | Thailand | II | 32 | 32 |
| Pi40 | CBS 777.84 | Mosquito larva | India | II | 64 | 16 |
| Pi38 | CBS 101039 | Human (eye) | India | II | 32 | 16 |
| Pi39 | CBS 702.83 | Equine | Japan | II | 32 | 8 |
| Pi37 | ATCC 28251 | Equine | Papua New Guinea | II | 64 | 16 |
| Pi51 | NAN06 | Environment | Thailand | III | 32 | 16 |
| Pi47 | SIMI 2921-45 | Human (eye) | Thailand | III | 16 | 16 |
| Pi49 | SIMI 7695-48 | Human (artery) | Thailand | III | 32 | 16 |
| Pi50 | ATCC 90586 | Human (skin) | USA | III | 32 | 16 |
The strain/reference identification numbers (ID), the hosts (tissues) and countries where the organisms were isolated, the rDNA-based genotypes (i.e., clades I, II, and III), and the disulfiram MICs determined by agar and broth susceptibility methods are indicated.
FIG 1.
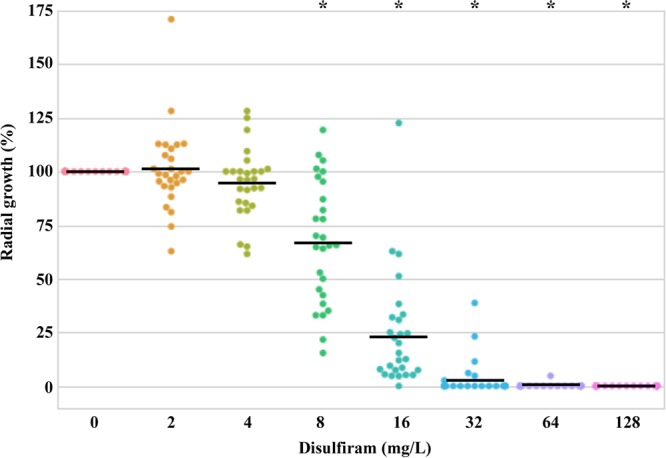
Agar-based susceptibility of P. insidiosum against disulfiram. The radial growths (%) of the organism (27 strains) after exposure to various concentrations of disulfiram (range, 0 to 128 mg/liter) are plotted. A straight line indicates mean growth. An asterisk represents significant growth reduction in relation to the no-drug control.
Identification of putative disulfiram targets in P. insidiosum.
The known target proteins of disulfiram (i.e., ALDH, DBH, CK, and URE) served as the query sequences and were subjected to a BLAST search against the genome-derived proteome of P. insidiosum (14,962 proteins) (27, 28). The DBH and CK sequences failed to match any P. insidiosum proteins. The URE sequence matched one protein, PINS00920015C (E value, 0.0; identity, 62%; similarity, 75%) (Table 2). The ALDH sequence matched 11 different proteins (Table 2), and among them PINS02440013A showed the best BLAST scores (E value, 3E–120; identity, 43%; similarity, 60%). All 12 matched proteins showed significant BLAST hits against the P. insidiosum transcriptome. Five proteins (i.e., PINS00920015C, PINS00560032A, PINS00450059A, PINS00850003A, and PINS00030096) can be mapped with 13 to 40 peptides of the P. insidiosum mascot proteome (Table 2).
TABLE 2.
Putative target proteins of disulfiram identified in the genome-derived proteome of Pythium insidiosuma
| Target proteins of disulfiram | Accession no. of query protein sequence | BLASTP against P. insidiosum proteome |
TBLASTN against transcriptome (E value) | No. of mapped peptide(s) in mascot library | ||
|---|---|---|---|---|---|---|
| Matched protein ID | E value | Accession | ||||
| Dopamine beta-hydroxylase (DBH) | P09172 | No match | ||||
| Carbamate kinase (CK) | 4OLC | No match | ||||
| Urease (URE) | 4H9M | PINS00920015C | 0.00 | GAX98700.1 | 0.00 | 40 |
| Aldehyde dehydrogenase (ALDH2) | P05091 | PINS02440013A | 3E–120 | GAY03418.1 | 0.00 | No match |
| PINS00560032A | 4E–66 | GAX96905.1 | 0.00 | 13 | ||
| PINS00240041A | 7E–66 | GAX94676.1 | 5E–179 | No match | ||
| PINS00460028A | 7E–61 | GAX96345.1 | 0.00 | No match | ||
| PINS00450059A | 2E–51 | GAX96237.1 | 0.00 | 21 | ||
| PINS00090050A | 1E–43 | GAX93307.1 | 0.00 | No match | ||
| PINS00450060A | 4E–43 | GAX96236.1 | 0.00 | No match | ||
| PINS00530007C | 6E–30 | GAX96738.1 | 0.00 | No match | ||
| PINS01590003A | 1E–22 | GAY01314.1 | 0.00 | No match | ||
| PINS00850003A | 5E–22 | GAX98398.1 | 0.00 | 14 | ||
| PINS00030096A | 5E–09 | GAX92574.1 | 0.00 | 18 | ||
The query protein sequences are deposited in the PDB (i.e., CK and URE) or UniProt (i.e., DBH and ALDH2) database. The transcriptome and in-house mascot proteome of P. insidiosum were used to validate the matched proteins.
Homology modeling of the target proteins and molecular docking of disulfiram.
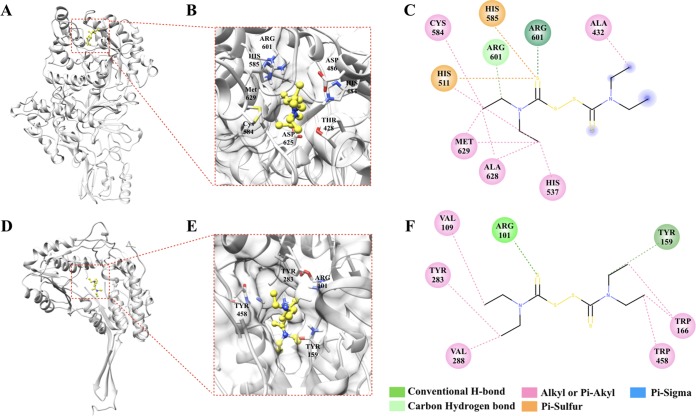
The best matched P. insidiosum proteins for URE (PINS00920015C) and ALDH (PINS02440013A) were subjected to homology modeling, using the Jack bean URE (PDB ID 3LA4) and the Pseudomonas γ-hydroxymuconic semialdehyde dehydrogenase (PDB ID 4GO4) as the prototypic templates, respectively. Disulfiram can be docked into the binding pockets of the P. insidiosum URE (amino acid residues: ALA432, HIS511, HIS537, CYS584, HIS585, ARG601, ALA628, and MET629), with a binding affinity of –4.0 Kcal/mol, and ALDH (amino acid residues: ARG101, VAL109, TYR159, TRP166, TYR283, VAL288, and TRP458) with a binding affinity of –6.1 Kcal/mol (Fig. 2). The most poses of docking could dock into the same binding pockets. The disulfiram-derived metabolites diethyldithiocarbamic acid (DDC), diethylamine (DEA), and diethyldithiomethylcarbamate (Me-DDC) can also be docked into the binding pocket of the P. insidiosum URE with the binding affinities of –3.4, –2.8, and –3.7 Kcal/mol, respectively (see Fig. S1 to S3 in the supplemental material). Likewise, these metabolites can be docked into the binding pocket of the P. insidiosum ALDH with binding affinities of –3.4, –2.8, and –3.6 Kcal/mol, respectively (see Fig. S1 to S3 in the supplemental material). Drug-target interactions (i.e., conventional H-bond, carbon hydrogen bond, alkyl or Pi-akyl bond, Pi-sulfur bond, Pi-Sigma bond, and Vander Waals bond) of the tested chemicals and the specific amino acid residues of the P. insidiosum URE and ALDH are shown in Fig. 2 (disulfiram), as well as Fig. S1 (DDC), S2 (DEA), and S3 (Me-DDC) in the supplemental material.
FIG 2.
Complexes of the ligand “disulfiram” and the predicted 3D structures of the urease (A to C) and aldehyde dehydrogenase (D to F) of P. insidiosum. Panels A and D show ribbon structure models, while panels B and E depict enlarged structure showing molecular docking of disulfiram. Panels C and F show the 2D interactions of disulfiram and the predicted binding sites of urease and aldehyde dehydrogenase, respectively. The interacting amino acids and types are shown in figures B, C, E, and F. The structures and complex interaction images were generated using the UCSF Chimera and Discovery Studio Visualizer (see Materials and Methods).
Disulfiram inhibits the URE and ALDH activities.
To demonstrate whether disulfiram inhibits the activity of URE, the mixtures of the Jack bean URE (concentration, 0.1 mg/ml) and the disulfiram (concentrations, 0 or 128 mg/liter) were assessed for biochemical activity, using the URE test tube assay. The assay was read positive in the absence of disulfiram (the agar color turned pink; Fig. 3B) but read negative in the presence of the drug (the agar color remained yellow; Fig. 3C). Likewise, to determine whether disulfiram can inactivate ALDH, the mixtures of the recombinant human ALDH2 (concentration, 0.1 mg/ml) and the disulfiram (concentrations, 0 or 128 mg/liter) were tested for the ALDH activity using a colorimetric assay. In the absence of disulfiram, the ALDH activity was 443.2 nmol of NADH/min/ml, whereas, in the presence of the drug, the ALDH activity was 6.8 nmol of NADH/min/ml.
FIG 3.
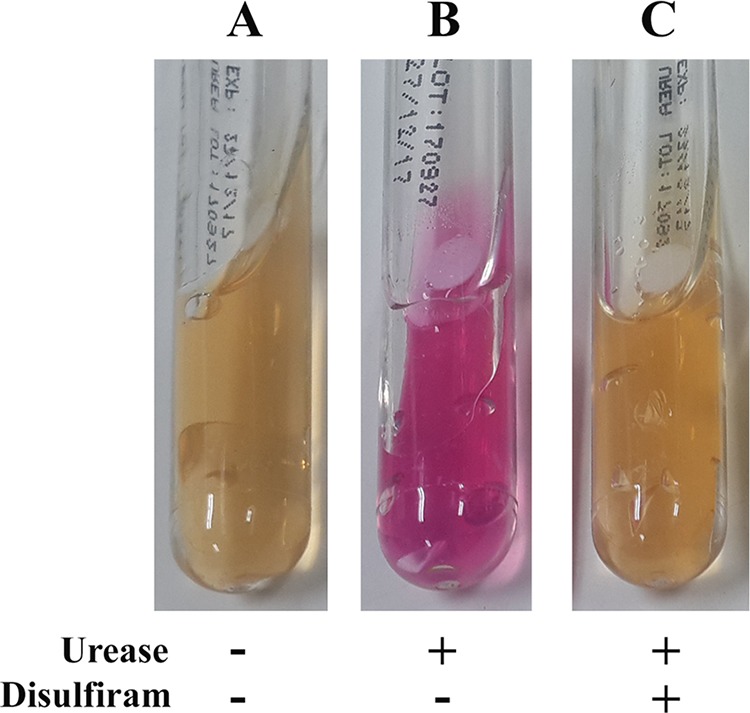
Biochemical effect of disulfiram on urease activity. The urease test tube assays were tested against DMSO (a diluent of disulfiram; this served as a control) (A), Jack bean urease alone (0.1 mg/ml) (B), and a preincubated mixture of Jack bean urease (0.1 mg/ml) and disulfiram (128 mg/liter) (C).
DISCUSSION
No standardized drug susceptibility method has been established for P. insidiosum. However, some investigators perform the microdilution-based susceptibility testing of P. insidiosum by adapting the CLSI M38-A2 method and using the inocula prepared from zoospores (asexual stage of the organism) (30, 31). In our hands, production of the zoospores is time-consuming and usually provides inadequate yield for the subsequent drug susceptibility assays. Alternatively, Brown et al. (32) and Lerksuthirat et al. (12) use the inocula prepared from agar plugs with actively growing P. insidiosum hyphae and performed the drug susceptibility assay by measuring the radial growth of the organism on a drug-containing agar plate. Their agar-based susceptibility method is easy, robust, and reproducible. For these reasons, we adopted the agar-based method for in vitro susceptibility testing of P. insidiosum against disulfiram. We also performed a broth-based susceptibility assay, which is a modified version of the agar-based susceptibility method that uses drug-containing broth rather than agar.
Pythiosis has high rates of morbidity and mortality. Most conventional antifungal drugs show unfavorable response against P. insidiosum. Drug repurposing (15, 16) could be a strategy to identify a potential FDA-approved drug for the control of P. insidiosum. Disulfiram (originally approved to treat alcoholism) exhibits antimicrobial activity against various pathogens (18–23). We explored whether disulfiram possesses any anti-P. insidiosum effect. As shown here, disulfiram inhibited the growths of all 27 P. insidiosum strains isolated from various hosts and geographic areas, in a dose-dependent manner (Table 1 and Fig. 1). The MIC range (8 to 32 mg/liter), the MIC50 (16 mg/liter), and the MIC90 (32 mg/liter) of the broth-based susceptibility method were lower than the MIC range (16 to 128 mg/liter), the MIC50 (32 mg/liter), and the MIC90 (64 mg/liter) of the agar-based susceptibility method (Table 1). In the broth-based method, P. insidiosum was completely immersed in disulfiram-containing liquid, whereas, in the agar-based method, the organism exposed the drug only on one side of the colony. The extent of drug exposure could explain the relatively lower MIC values of the broth-based method. Compared to other microorganisms, the MIC range of disulfiram against P. insidiosum (8 to 32 mg/liter [broth-based method]) was in line with that of the antibacterial drug-resistant S. aureus (8 to 32 mg/liter) (19) and the nontuberculous mycobacteria (32 mg/liter) (23) but markedly higher than that of M. tuberculosis (1.56 mg/liter) (22). Thus, disulfiram has an extended antimicrobial spectrum that covers the oomycete microorganisms, including P. insidiosum.
In an attempt of measuring disulfiram in blood, Brown et al. reported that 12 h after a single 200-mg dose, the blood level of the drug was approximately 4.5 mg/liter (33). Moreover, after oral administration of the daily dose of 200 mg for 14 days, the blood level of disulfiram increased to 8.3 mg/liter (34, 35). Since the recommended daily dose of disulfiram could be optimized to 500 mg (36), it is possible that the blood concentration of the drug can reach a level that potentially inhibits the susceptible organisms (i.e., P. insidiosum) that have an MIC50 of >8 mg/liter. Regarding toxicity, disulfiram has a 50% lethal dose (LD50) of 8.6 g/kg (orally in rats [https://www.drugbank.ca/]), and thus it is a practically nontoxic drug, based on the Hodge and Sterner criteria (https://www.ccohs.ca/oshanswers/chemicals/ld50.html). However, a future susceptibility experiment in an animal model of pythiosis should provide insightful information on in vivo anti-P. insidiosum effect and toxicity of disulfiram.
In other organisms, inactivation of the enzyme ALDH, URE, CK, or DBH contributes to the chemotherapeutic or antimicrobial activities of disulfiram (21, 24–26). We initially assessed these targets of disulfiram in P. insidiosum. The BLAST search only identified ALDH (11 copies) and URE (one copy), but not CK and DBH, in the genome-derived proteome of P. insidiosum (27, 28) (Table 2). Interestingly, ALDH and URE are considered a microbial virulence determinant or essential enzyme that can promote an infection (37–40), suggesting that these proteins are potential drug targets. By homology modeling and molecular docking, disulfiram can bind the putative ALDH and URE of P. insidiosum at the relatively low energies of –6.1 and –4.0 Kcal/mol, respectively (Fig. 2). In addition, sequence homology analysis revealed that the P. insidiosum URE contains “VCHHL” (amino acid residues 583 to 587), which is a conserved region found in all bacterial, plant, and fungal UREs and plays a crucial role in URE function (24). Disulfiram can form protein-ligand interactions at this conserved region of the P. insidiosum URE, covering the amino acid residues CYS584 and HIS585 (Fig. 2), which suggests a potential anti-URE mechanism of the drug in this organism.
The human gastrointestinal tract absorbs at least 80% of the ingested disulfiram into the blood circulation, where the drug can transform to DDC by the endogenous thiols and the glutathione reductase of red blood cells (41). The DDC then forms CS2, DEA, Me-DDC, and other metabolites (41). Disulfiram and its metabolites circulate and distribute to various tissues, such as the liver, kidney, heart, lung, brain, testes, spleen, and intestine (42). Drug metabolism is a clinical concern for the repurposed use of disulfiram in cancer, as it may decrease the effective form and lead to an unfavorable response of the drug against cancer cells (43). Metabolism of disulfiram is also a concern for infectious diseases, since the formation of various metabolites might compromise the antimicrobial activities of the drug. To address this issue, Chen et al. proposed the use of disulfiram-loaded lipid emulsion, which minimizes degradation and improves the chemical stability of the drug in the blood (43). However, since the inhibitory property of the disulfiram-derived metabolites against P. insidiosum is concerned, we performed the molecular docking and demonstrated that DDC, DEA, and Me-DDC are capable of binding P. insidiosum URE and ALDH at low energies of less than –2.8 Kcal/mol (Fig. S1 to S3). In agreement with the docking analysis, disulfiram inhibited the biochemical activities of both ALDH (represented by the recombinant human ALDH2) and URE (represented by Jack bean URE). Taken together, disulfiram and its metabolites can bind and inactivate multiple proteins of P. insidiosum (i.e., ALDH and URE), that may contribute to the broad-spectrum antimicrobial property of this FDA-approved chemical.
Not only does P. insidiosum cause systemic infection, but it also causes a serious local infection of the eye (2). Topical administration of disulfiram (which has limited water solubility) is possible in the treatment of ocular pythiosis because an ophthalmic formulation of the drug can be prepared at a high concentration (i.e., 0.5% [vol/wt]) by using solid nanoparticles (44). Such ophthalmic formulation shows good corneal penetration and undetectable toxicity in the treatment of glaucoma in a rabbit model (44). In contrast to the systemic infection, metabolism or decomposition of disulfiram should be less concerned in the ocular infection due to a lower chance of the drug to expose the glutathione reductase system of red blood cells.
Deferasirox (originally approved to treat chronic iron overload) has been also repurposed for the control of P. insidiosum (30). The MIC range of deferasirox against P. insidiosum (12.5 to 50 mg/liter) is slightly higher than that of disulfiram (8 to 32 mg/liter). Micafungin, when combined with deferasirox, as opposed to being tested alone, shows a synergistic antimicrobial effect in most P. insidiosum strains tested (45). Likewise, vancomycin, when combined with disulfiram, shows an enhanced antimicrobial activity against S. aureus (19). A combination of disulfiram and other drugs might provide a synergistic anti-P. insidiosum effect, which requires a further study.
In conclusion, disulfiram (approved for the alcoholism treatment) inhibits the growth of many pathogenic microorganisms, including P. insidiosum, by inactivating several proteins, i.e., ALDH, URE, CK, or DBH (21, 24–26). Various strains of P. insidiosum from different hosts and geographic origins were susceptible to disulfiram in a dose-dependent manner and at MICs similar to those for other pathogens. ALDH and URE are present in P. insidiosum and serve as potential binding targets of disulfiram. Evaluation of disulfiram in an animal model of pythiosis could provide more information on the in vivo drug efficiency against P. insidiosum. Disulfiram is an FDA-approved drug that could be repurposed as a new treatment option for pythiosis.
MATERIALS AND METHODS
Microorganisms.
Twenty-seven strains of P. insidiosum, isolated from different hosts and geographic areas (Table 1), were recruited for the in vitro susceptibility test against disulfiram. Identities of the organisms were verified by rDNA sequence analysis. All strains were maintained on Sabouraud dextrose (SD) agar (pH 7.2) and subcultured once a month until use.
In vitro susceptibility assays.
The growths of 27 P. insidiosum strains (Table 1), upon exposure to disulfiram, were measured using the agar-based (12, 32) and broth-based (see below) susceptibility methods. The experiments were performed in duplicate. SD agar plugs (5 mm in diameter) with actively growing mycelium were excised from a P. insidiosum colony (∼7 days of age) and served as the inocula for the susceptibility assays. An agar plug with the organism was placed on the SD agar or immersed in the SD broth containing various concentrations (i.e., 0, 2, 4, 8, 16, 32, 64, and 128 mg/liter) of disulfiram (purity, ≥98%; solvent, dimethyl sulfoxide [DMSO]; manufacturer, Unidrug Innovative Pharma Technologies, Ltd., India) and incubated at 37°C for 2 days. For the agar-based susceptibility method (12, 32), two perpendicular diameters of a P. insidiosum colony were averaged, subtracted by the agar plug diameter (5 mm), and divided by half. The actual radial growth of a disulfiram-exposed strain was calculated to the “percent growth” in relation to that of the same strain without drug exposure (control). The percent growths of all P. insidiosum strains tested with different disulfiram concentrations were compared, using the Stata version 10 (StataCorp LLC) and a paired t test. Dot plots representing the percent growths of P. insidiosum was generated, using the Python program version 3.7.1 (https://www.python.org) and a modified script from Seaborn (https://seaborn.pydata.org). For the broth susceptibility method (which is a modified version of the agar-based susceptibility method that uses drug-containing broth rather than agar), an agar plug with a growing organism was immersed in a test tube containing 3 ml of SD broth with or without disulfiram. After incubation at 37°C for 2 days, each strain was checked for growth by naked eyes and reported as susceptible (if both replicates showed no growth) or resistant (if at least one replicate grew).
Proteogenomic analysis of putative targets of disulfiram.
The protein sequences of ALDH (UniProt ID P05091) and DBH (UniProt ID P09172) were retrieved from the UniProt database (https://www.uniprot.org), whereas those of CK (PDB ID 4OLC) and URE (PDB ID 4H9M) were retrieved from the Protein Data Bank (https://www.rcsb.org). To identify homology sequences, the retrieved proteins were BLASTP searched against the P. insidiosum genome-derived proteome (27, 28), using the locally installed blast 2.2.28+ program (https://www.ncbi.nlm.nih.gov; E value cutoff, <10−6). The in-house mascot proteomic library and transcriptome of P. insidiosum were used to validate the matched proteins and their encoding genes, as described previously (28, 46).
Homology modeling of P. insidiosum proteins.
The X-ray crystallography-derived three-dimensional (3D) structures of two proteins, the Jack bean URE of Canavalia ensiformis (PDB ID 3LA4) and the γ-hydroxymuconic semialdehyde dehydrogenase of Pseudomonas species strain WBC-3 (PDB ID 4GO4), were retrieved from the Protein Data Bank. These 3D structures were used as protein templates for generating 3D structures of the URE (accession no. BBE00775.1) and the betaine ALDH (accession no. GAY03418.1) of P. insidiosum, respectively. Binding pockets of the obtained 3D structures were predicted using the I-TASSER Suite (47). The UCSF Chimera program, v1.10.2 (http://www.cgl.ucsf.edu/chimera/) (48), was used to visualize the predicted 3D structures of the P. insidiosum proteins.
Molecular docking.
The 3D structures of disulfiram (PubChem ID 3117), DDC (PubChem ID 8987), and DEA (PubChem ID 8021) were downloaded from the PubChem server (http://pubchem.ncbi.nlm.nih.gov/). The 3D structure of Me-DDC was constructed based on the DDC structure using the PreADMET server (https://preadmet.bmdrc.kr/introduction/). Molecular docking analyses of these chemical 3D structures (i.e., disulfiram, DDC, DEA, and Me-DDC) and the P. insidiosum proteins (i.e., URE and betaine ALDH) were performed using the AutoDock Vina program (49) and the PyRx suite open-source software version 0.9.7, which is a knowledge-based empirical scoring function of a ligand-protein complex (50). The input files were converted to the pdbqt file format by the AutoDock Vina program. The grid center (X:Y:Z) and dimension (X:Y:Z) of URE were 90:52:84 and 40:35:35, respectively, while those of ALDH were 75:80:76 and 22:23:25. The docking analyses were relied on the Lamarckian generic algorithm (GA) using the following default parameters: 20 GA runs, 150 individuals in the population, 270,000 maximum numbers of energy evaluation, and a 0.02 gene mutation rate and a 0.8 crossover rate. The outputs of 10 best binding poses for each docking run were stored for further evaluation of protein-ligand interaction, using the Discovery Studio Visualizer version 17.2.0 (Dassault Systemes Biovia Corp.).
Inhibitory activities of disulfiram.
Inhibitory activities of disulfiram against the known URE and ALDH were evaluated. The Jack bean URE (Sigma-Aldrich) (final concentration, 0.1 mg/ml) and disulfiram (final concentrations, 0 or 128 mg/liter) were mixed at the final volume of 30 μl and incubated at room temperature for 10 min. The mixture was then added onto a 6-mm-diameter grade-AA disc paper (Whatman), already placed in a URE test tube (CLINAG, Thailand). After incubation at 37°C for 7 days, the URE test was read as positive if the agar color was changed to pink or as negative if the agar color remained unchanged (yellow). The recombinant human ALDH2 (Sigma-Aldrich) (final concentration, 10 μg/ml) and disulfiram (final concentrations, 0 or 128 mg/liter) were mixed at the final volume of 50 μl and incubated at room temperature for 10 min. The ALDH activity (reported as the nmol of NADH/min/ml) of the mixture was measured by using an ALDH activity colorimetric assay kit (Sigma-Aldrich).
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the financial support provided by Mahidol University (grant ngor-por 54/2561 [T.K.]); Thailand Research Fund (grants RSA6280092 and BRG5980009 [T.K.] and MRG6080067 [S.T.]), the Faculty of Medicine, Ramathibodi Hospital, Mahidol University (grant CF_61007 [T.K.]); and King Mongkut’s University of Technology Thonburi through the KMUTT 55th Anniversary Commemorative Fund (W.K.).
We thank Pachara Pokavipanun and Supitchaya Pantia for research assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00609-19.
REFERENCES
- 1.Thianprasit M, Chaiprasert A, Imwidthaya P. 1996. Human pythiosis. Curr Top Med Mycol 7:43–54. [PubMed] [Google Scholar]
- 2.Krajaejun T, Sathapatayavongs B, Pracharktam R, Nitiyanant P, Leelachaikul P, Wanachiwanawin W, Chaiprasert A, Assanasen P, Saipetch M, Mootsikapun P, Chetchotisakd P, Lekhakula A, Mitarnun W, Kalnauwakul S, Supparatpinyo K, Chaiwarith R, Chiewchanvit S, Tananuvat N, Srisiri S, Suankratay C, Kulwichit W, Wongsaisuwan M, Somkaew S. 2006. Clinical and epidemiological analyses of human pythiosis in Thailand. Clin Infect Dis 43:569–576. doi: 10.1086/506353. [DOI] [PubMed] [Google Scholar]
- 3.Gaastra W, Lipman LJA, De Cock A, Exel TK, Pegge RBG, Scheurwater J, Vilela R, Mendoza L. 2010. Pythium insidiosum: an overview. Vet Microbiol 146:1–16. doi: 10.1016/j.vetmic.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 4.Chareonsirisuthigul T, Khositnithikul R, Intaramat A, Inkomlue R, Sriwanichrak K, Piromsontikorn S, Kitiwanwanich S, Lowhnoo T, Yingyong W, Chaiprasert A, Banyong R, Ratanabanangkoon K, Brandhorst TT, Krajaejun T. 2013. Performance comparison of immunodiffusion, enzyme-linked immunosorbent assay, immunochromatography, and hemagglutination for serodiagnosis of human pythiosis. Diagn Microbiol Infect Dis 76:42–45. doi: 10.1016/j.diagmicrobio.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 5.Jindayok T, Piromsontikorn S, Srimuang S, Khupulsup K, Krajaejun T. 2009. Hemagglutination test for rapid serodiagnosis of human pythiosis. Clin Vaccine Immunol 16:1047–1051. doi: 10.1128/CVI.00113-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inkomlue R, Larbcharoensub N, Karnsombut P, Lerksuthirat T, Aroonroch R, Lohnoo T, Yingyong W, Santanirand P, Sansopha L, Krajaejun T. 2016. Development of an anti-elicitin antibody-based immunohistochemical assay for diagnosis of pythiosis. J Clin Microbiol 54:43–48. doi: 10.1128/JCM.02113-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keeratijarut A, Lohnoo T, Yingyong W, Rujirawat T, Srichunrusami C, Onpeaw P, Chongtrakool P, Brandhorst TT, Krajaejun T. 2015. Detection of the oomycete Pythium insidiosum by real-time PCR targeting the gene coding for exo-1,3-β-glucanase. J Med Microbiol 64:971–977. doi: 10.1099/jmm.0.000117. [DOI] [PubMed] [Google Scholar]
- 8.Keeratijarut A, Lohnoo T, Yingyong W, Nampoon U, Lerksuthirat T, Onpaew P, Chongtrakool P, Krajaejun T. 2014. PCR amplification of a putative gene for exo-1,3-β-glucanase to identify the pathogenic oomycete Pythium insidiosum. Asian Biomed 8:637–644. doi: 10.5372/1905-7415.0805.338. [DOI] [Google Scholar]
- 9.Intaramat A, Sornprachum T, Chantrathonkul B, Chaisuriya P, Lohnoo T, Yingyong W, Jongruja N, Kumsang Y, Sandee A, Chaiprasert A, Banyong R, Santurio JM, Grooters AM, Ratanabanangkoon K, Krajaejun T. 2016. Protein A/G-based immunochromatographic test for serodiagnosis of pythiosis in human and animal subjects from Asia and Americas. Med Mycol 54:641–647. doi: 10.1093/mmy/myw018. [DOI] [PubMed] [Google Scholar]
- 10.Grooters AM, Gee MK. 2002. Development of a nested polymerase chain reaction assay for the detection and identification of Pythium insidiosum. J Vet Intern Med 16:147–152. doi:. [DOI] [PubMed] [Google Scholar]
- 11.Keeratijarut A, Karnsombut P, Aroonroch R, Srimuang S, Sangruchi T, Sansopha L, Mootsikapun P, Larbcharoensub N, Krajaejun T. 2009. Evaluation of an in-house immunoperoxidase staining assay for histodiagnosis of human pythiosis. Southeast Asian J Trop Med Public Health 40:1298–1305. [PubMed] [Google Scholar]
- 12.Lerksuthirat T, Sangcakul A, Lohnoo T, Yingyong W, Rujirawat T, Krajaejun T. 2017. Evolution of the sterol biosynthetic pathway of Pythium insidiosum and related oomycetes contributes to antifungal drug resistance. Antimicrob Agents Chemother 61:e02352-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendoza L, Newton JC. 2005. Immunology and immunotherapy of the infections caused by Pythium insidiosum. Med Mycol 43:477–486. doi: 10.1080/13693780500279882. [DOI] [PubMed] [Google Scholar]
- 14.Krajaejun T, Pracharktam R, Wongwaisayawan S, Rochanawutinon M, Kunakorn M, Kunavisarut S. 2004. Ocular pythiosis: is it under-diagnosed? Am J Ophthalmol 137:370–372. doi: 10.1016/S0002-9394(03)00908-5. [DOI] [PubMed] [Google Scholar]
- 15.Zheng W, Sun W, Simeonov A. 2018. Drug repurposing screens and synergistic drug-combinations for infectious diseases. Br J Pharmacol 175:181–191. doi: 10.1111/bph.13895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburn TT, Thor KB. 2004. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 17.Shim JS, Liu JO. 2014. Recent advances in drug repositioning for the discovery of new anticancer drugs. Int J Biol Sci 10:654–663. doi: 10.7150/ijbs.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis PM, Dronsfield AT. 2013. Antabuse’s diamond anniversary: still sparkling on? Drug Alcohol Rev 32:342–344. doi: 10.1111/dar.12018. [DOI] [PubMed] [Google Scholar]
- 19.Long TE. 2017. Repurposing thiram and disulfiram as antibacterial agents for multidrug-resistant Staphylococcus aureus infections. Antimicrob Agents Chemother 61:e00898-17. doi: 10.1128/AAC.00898-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velasco-García R, Chacón-Aguilar VM, Hervert-Hernández D, Muñoz-Clares RA. 2003. Inactivation of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa and Amaranthus hypochondriacus L. leaves by disulfiram. Chem Biol Interact 143–144:149–158. doi: 10.1016/S0009-2797(02)00199-0. [DOI] [PubMed] [Google Scholar]
- 21.Galkin A, Kulakova L, Lim K, Chen CZ, Zheng W, Turko IV, Herzberg O. 2014. Structural basis for inactivation of Giardia lamblia carbamate kinase by disulfiram. J Biol Chem 289:10502–10509. doi: 10.1074/jbc.M114.553123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horita Y, Takii T, Yagi T, Ogawa K, Fujiwara N, Inagaki E, Kremer L, Sato Y, Kuroishi R, Lee Y, Makino T, Mizukami H, Hasegawa T, Yamamoto R, Onozaki K. 2012. Antitubercular activity of disulfiram, an antialcoholism drug, against multidrug- and extensively drug-resistant Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother 56:4140–4145. doi: 10.1128/AAC.06445-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das S, Garg T, Chopra S, Dasgupta A. 2019. Repurposing disulfiram to target infections caused by non-tuberculous mycobacteria. J Antimicrob Chemother 74:1317–1322. doi: 10.1093/jac/dkz018. [DOI] [PubMed] [Google Scholar]
- 24.Díaz-Sánchez ÁG, Alvarez-Parrilla E, Martínez-Martínez A, Aguirre-Reyes L, Orozpe-Olvera JA, Ramos-Soto MA, Núñez-Gastélum JA, Alvarado-Tenorio B, de la Rosa LA. 2016. Inhibition of urease by disulfiram, an FDA-approved thiol reagent used in humans. Molecules 21:1628. doi: 10.3390/molecules21121628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, Pon A, Knox C, Wilson M. 2018. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res 46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaval-Cruz M, Weinshenker D. 2009. Mechanisms of disulfiram-induced cocaine abstinence: antabuse and cocaine relapse. Mol Interv 9:175–187. doi: 10.1124/mi.9.4.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rujirawat T, Patumcharoenpol P, Lohnoo T, Yingyong W, Lerksuthirat T, Tangphatsornruang S, Suriyaphol P, Grenville-Briggs LJ, Garg G, Kittichotirat W, Krajaejun T. 2015. Draft genome sequence of the pathogenic oomycete Pythium insidiosum strain Pi-S, isolated from a patient with pythiosis. Genome Announc 3:e00574-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rujirawat T, Patumcharoenpol P, Lohnoo T, Yingyong W, Kumsang Y, Payattikul P, Tangphatsornruang S, Suriyaphol P, Reamtong O, Garg G, Kittichotirat W, Krajaejun T. 2018. Probing the phylogenomics and putative pathogenicity genes of Pythium insidiosum by oomycete genome analyses. Sci Rep 8:4135. doi: 10.1038/s41598-018-22540-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rujirawat T, Sridapan T, Lohnoo T, Yingyong W, Kumsang Y, Sae-Chew P, Tonpitak W, Krajaejun T. 2017. Single nucleotide polymorphism-based multiplex PCR for identification and genotyping of the oomycete Pythium insidiosum from humans, animals and the environment. Infect Genet Evol 54:429–436. doi: 10.1016/j.meegid.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Zanette RA, Alves SH, Pilotto MB, Weiblen C, Fighera RA, Wolkmer P, Flores MM, Santurio JM. 2013. Iron chelation therapy as a treatment for Pythium insidiosum in an animal model. J Antimicrob Chemother 68:1144–1147. doi: 10.1093/jac/dks534. [DOI] [PubMed] [Google Scholar]
- 31.Loreto ES, Tondolo JSM, Oliveira DC, Santurio JM, Alves SH. 2018. In vitro activities of miltefosine and antibacterial agents from the macrolide, oxazolidinone, and pleuromutilin classes against Pythium insidiosum and Pythium aphanidermatum. Antimicrob Agents Chemother 62:e01678-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown TA, Grooters AM, Hosgood GL. 2008. In vitro susceptibility of Pythium insidiosum and a Lagenidium sp to itraconazole, posaconazole, voriconazole, terbinafine, caspofungin, and mefenoxam. Am J Vet Res 69:1463–1468. doi: 10.2460/ajvr.69.11.1463. [DOI] [PubMed] [Google Scholar]
- 33.Brown MW, Porter GS, Williams AE. 1974. Proceedings: the determination of disulfiram in blood, and of exhaled carbon disulphide using cathode ray polarography. J Pharm Pharmacol 26(Suppl):95P–96P. doi: 10.1111/j.2042-7158.1974.tb10123.x. [DOI] [PubMed] [Google Scholar]
- 34.Malcolm MT, Madden JS, Williams AE. 1974. Disulfiram implantation critically evaluated. Br J Psychiatry 125:485–489. doi: 10.1192/bjp.125.5.485. [DOI] [PubMed] [Google Scholar]
- 35.Malcolm MT. 1977. Disulfiram blood levels. Br Med J 2:457. doi: 10.1136/bmj.2.6084.457-d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright C, Moore RD. 1990. Disulfiram treatment of alcoholism. Am J Med 88:647–655. doi: 10.1016/0002-9343(90)90534-K. [DOI] [PubMed] [Google Scholar]
- 37.Velasco-García R, Zaldívar-Machorro VJ, Mújica-Jiménez C, González-Segura L, Muñoz-Clares RA. 2006. Disulfiram irreversibly aggregates betaine aldehyde dehydrogenase: a potential target for antimicrobial agents against Pseudomonas aeruginosa. Biochem Biophys Res Commun 341:408–415. doi: 10.1016/j.bbrc.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 38.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. 2000. Urease as a virulence factor in experimental cryptococcosis. Infect Immun 68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutherford JC. 2014. The emerging role of urease as a general microbial virulence factor. PLoS Pathog 10:e1004062. doi: 10.1371/journal.ppat.1004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mora D, Arioli S. 2014. Microbial urease in health and disease. PLoS Pathog 10:e1004472. doi: 10.1371/journal.ppat.1004472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johansson B. 1992. A review of the pharmacokinetics and pharmacodynamics of disulfiram and its metabolites. Acta Psychiatr Scand Suppl 369:15–26. [DOI] [PubMed] [Google Scholar]
- 42.Stromme JH, Eldjarn L. 1966. Distribution and chemical forms of diethyldithiocarbamate and tetraethylthiuram disulphide (disculfiram) in mice in relation to radioprotection. Biochem Pharmacol 15:287–297. doi: 10.1016/0006-2952(66)90300-5. [DOI] [PubMed] [Google Scholar]
- 43.Chen X, Zhang L, Hu X, Lin X, Zhang Y, Tang X. 2015. Formulation and preparation of a stable intravenous disulfiram-loaded lipid emulsion. Eur J Lipid Sci Technol 117:869–878. doi: 10.1002/ejlt.201400278. [DOI] [Google Scholar]
- 44.Nagai N, Yoshioka C, Mano Y, Tnabe W, Ito Y, Okamoto N, Shimomura Y. 2015. A nanoparticle formulation of disulfiram prolongs corneal residence time of the drug and reduces intraocular pressure. Exp Eye Res 132:115–123. doi: 10.1016/j.exer.2015.01.022. [DOI] [PubMed] [Google Scholar]
- 45.Zanette RA, Jesus FPK, Pilotto MB, Weiblen C, Pötter L, Ferreiro L, Alves SH, Santurio JM. 2015. Micafungin alone and in combination therapy with deferasirox against Pythium insidiosum. J Mycol Med 25:91–94. doi: 10.1016/j.mycmed.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Krajaejun T, Lerksuthirat T, Garg G, Lowhnoo T, Yingyong W, Khositnithikul R, Tangphatsornruang S, Suriyaphol P, Ranganathan S, Sullivan TD. 2014. Transcriptome analysis reveals pathogenicity and evolutionary history of the pathogenic oomycete Pythium insidiosum. Fungal Biol 118:640–653. doi: 10.1016/j.funbio.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dallakyan S, Olson AJ. 2015. Small-molecule library screening by docking with PyRx. Methods Mol Biol 1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.