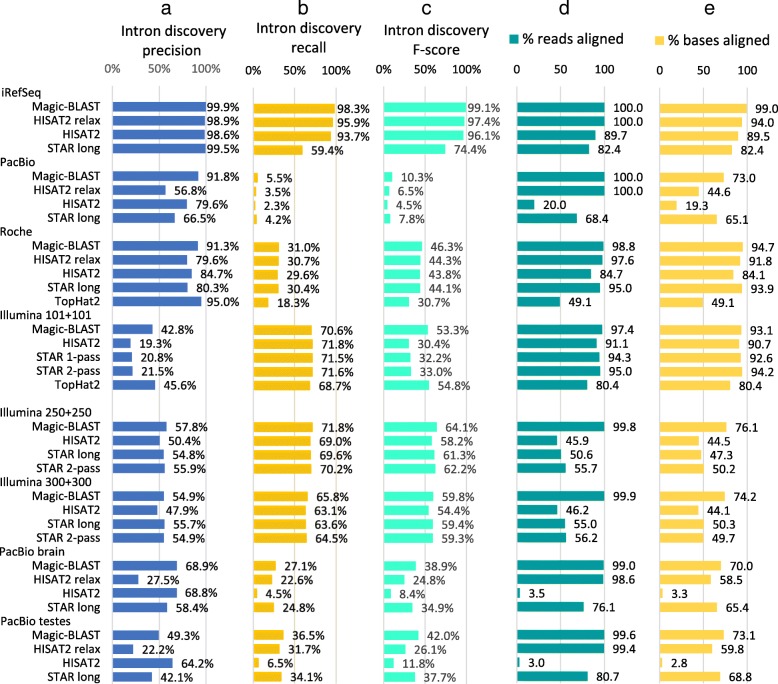
Fig. 5.
Performance of the aligners on intron discovery and alignment statistics measured on iRefSeq and seven experimental datasets. a Intron discovery precision p = (TP/TP + FP). b recall r = (TP/TP + FN) and c) F-score (2pr/p + r). All introns are counted, including those with a single support. An intron annotated in iRefSeq is counted as true, an unannotated intron as false. d percentage of reads aligned. e percentage of bases aligned. Average sequenced length per fragment are: iRefSeq 3427 bases, PacBio 1854 bases, Roche 348 bases, Illumina 202, 500 and 600 bases, PacBio 1300 and 1323 bases. The longer the reads, the more specific their alignments are