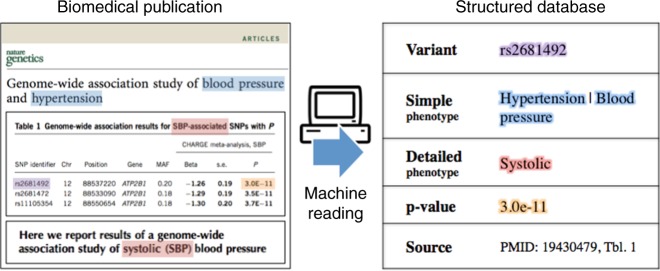
Fig. 1.
The automated information extraction system used to compile GWASkb. The GWASkb system takes as input a set of biomedical publications retrieved from PubMed Central (left) and automatically creates a structured database of GWAS associations described in these publications (right). For each association, the system identifies a genetic variant (purple), a high-level phenotype (pertaining to all variants in the publication), a detailed low-level phenotype (specific to individual variants, if available; red), and a p value (orange). Acronyms are also resolved (red)