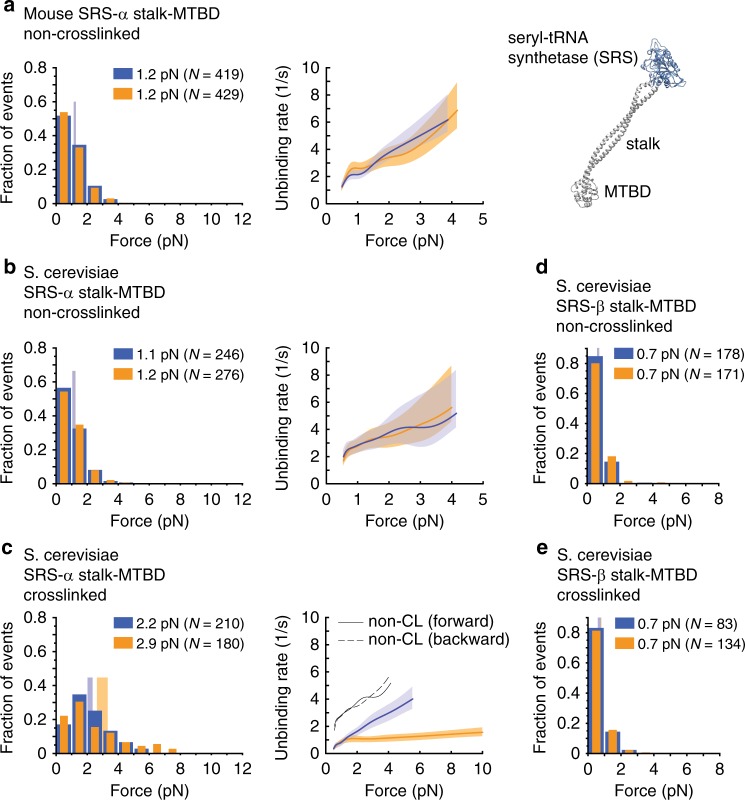
Fig. 4.
Unbinding-force behaviors of the SRS-stalk-MTBD constructs. a, left: Normalized histograms of primary forward and backward unbinding forces for the SRS construct with the mouse stalk helices (a.a. 3019–3309) fused in the non-cross-linked α-registry (SRS 85:82), with mean values noted (95% CIs [1.1, 1.3] and [1.1, 1.3] pN, estimated by bootstrapping 4,000 samples). a, middle: Unbinding rate vs. force derived from the data on the left. The shaded areas are 95% CIs for the mean rates, estimated by bootstrapping. a, right: Model of monomeric T. thermophilus seryl-tRNA synthetase (SRS) (based on PDB entry 3ERR) fused to the near-full-length stalk and MTBD of dynein in the β-registry of the stalk helices (generated by aligning PDB entry 3WUQ and 4RH7, see Supplementary Note 1). b As in (a) but for the SRS construct with the S. cerevisiae stalk helices (a.a. 3019–3309) fused in the non-cross-linked α-registry (95% CIs [1.0, 1.2] and [1.1, 1.2] pN). c As in (b) but with the stalk helices cross-linked (K3077C, A3250C) in the α-registration (95% CIs [2.0, 2.4] and [2.6, 3.3] pN). d As in (b) but for SRS fused to the S. cerevisiae stalk and MTBD in the weak MT-binding β-registry (SRS 89:82) (a.a. 3015–3309) (95% CIs [0.6, 0.7] and [0.6, 0.7] pN). e As in (d) but with the stalk helices cross-linked (I3076C, L3247C) in the β-registration (95% CIs [0.6, 0.7] and [0.7, 0.8] pN). Source data are provided as a Source Data file