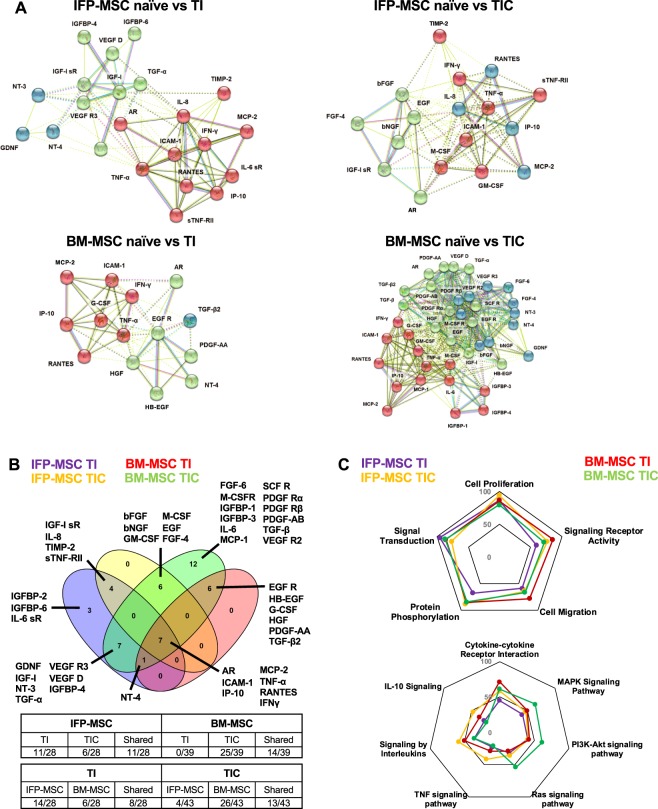
Figure 5.
Protein interactomes of IFP-MSC and BM-MSC secretory profiling pre- and post-TI and TIC priming. (A) STRING analysis of those proteins with statistical differences between naïve and TI-/TIC- priming conditions (total of 46) was performed using all available interaction sources and 0.4 as a confidence interaction score. K-means algorithm resulted into firm clustering of the differentially expressed factors into 3 groups (different colours) indicating high protein interaction within each group. (B) Venn diagram showing shared proteins among the groups, with AR, ICAM-1, IP-10, MCP-2, TNF-α, RANTES and IFN-γ present in both MSC types and in inflammatory and pro-fibrotic conditions. Table shows the number of proteins shared by different groups and conditions. (C) Biological processes and KEGG/reactome pathways analyses revealed different type and number of proteins affected post-TI and -TIC priming for both IFP-MSC and BM-MSC. Graph percentages represent the number of proteins involved in a specific pathway/function, related to the total amount of proteins differentially expressed between naïve and TI-/TIC-priming conditions.