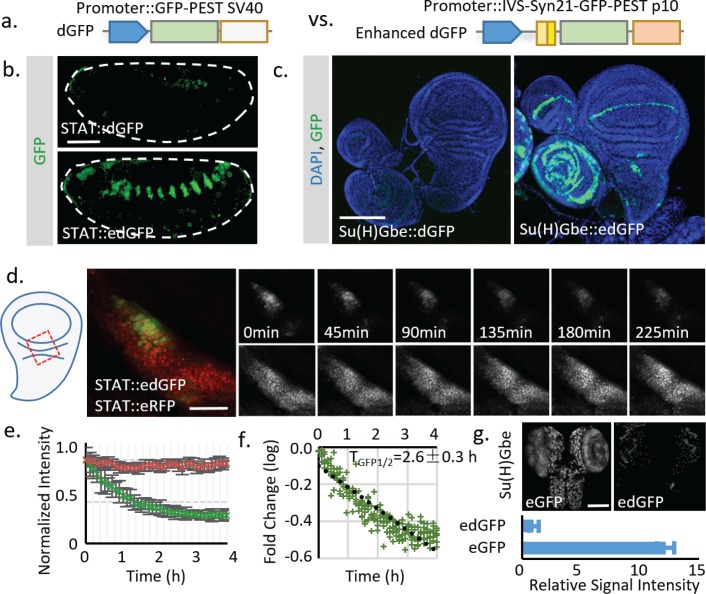
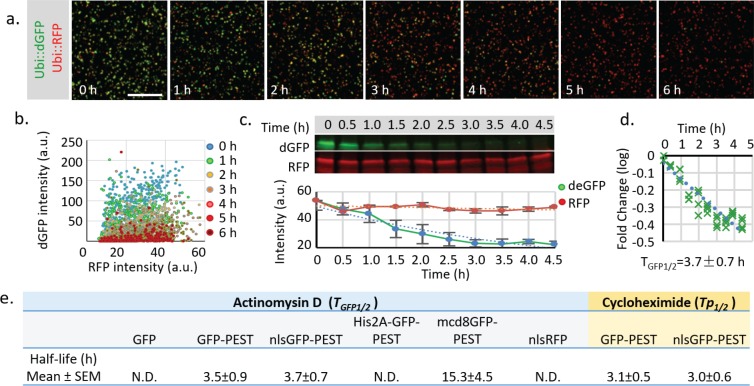
Figure 2. Using translational enhancers to increase the signal from destabilized fluorescent reporters for in vivo study.
(a) Illustration of the regular and destabilized GFP reporters. All the GFPs used in this study are the fast folding superfolder GFP (sfGFP). Destabilized GFP is labeled as dGFP, and dGFP reporter with translational enhancing elements is labeled as edGFP. All FPs used in this study contain the SV40 nuclear localization signal (NLS) at the N-terminus to facilitate signal segregation unless specified otherwise. (b, c) Comparison of dGFP and edGFP controlled by 6XSTAT response element in fly embryos and Su(H)Gbe Notch responding element in third instar wing imaginal discs. Images were taken with identical exposure. The contour of the embryo is outlined. (d) Dissected fly wing disc, expressing the STAT::edGFP and STAT:: eRFP, cultured ex vivo. Tissue was treated with 10 μM Actinomycin D to block transcription. STAT at the hinge region of the wing disc was imaged every 5 min for 4.5 hr. (e) The intensities of both dGFP and RFP were measured over time. Data from three independent replicates were collected and plotted. (f) The in vivo reporter half-life (TGFP1/2, representing effects of both Tm1/2 and Tp1/2) was estimated by linear regression of fluorescent intensity (in logarithmic scale). 95% confidence interval was calculated from linear regression. (g) Regular GFP and dGFP are expressed under Su(H)Gbe together with translational enhancers. Images were taken under identical parameters. The total fluorescent intensity from both reporters was plotted below with the intensity normalized to the dGFP signal. Data were collected from 10 different brains for each genotype. Scale bar: (b) 50 μm; (c, g) 100 μm; (d) 25 μm. Error bar: s.e.m.