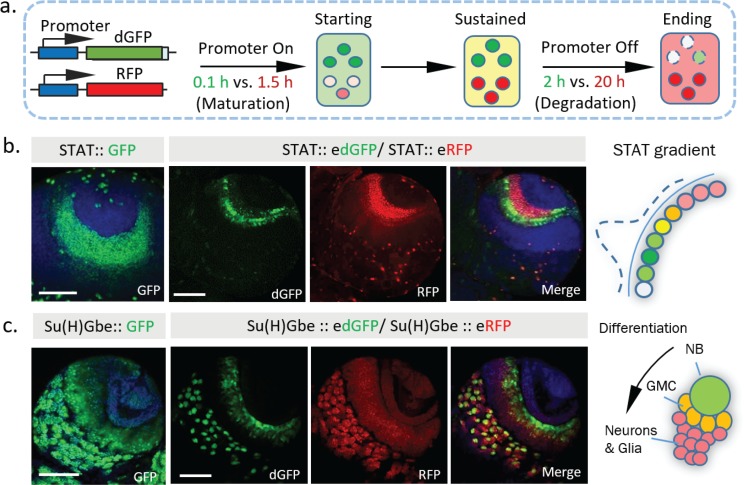
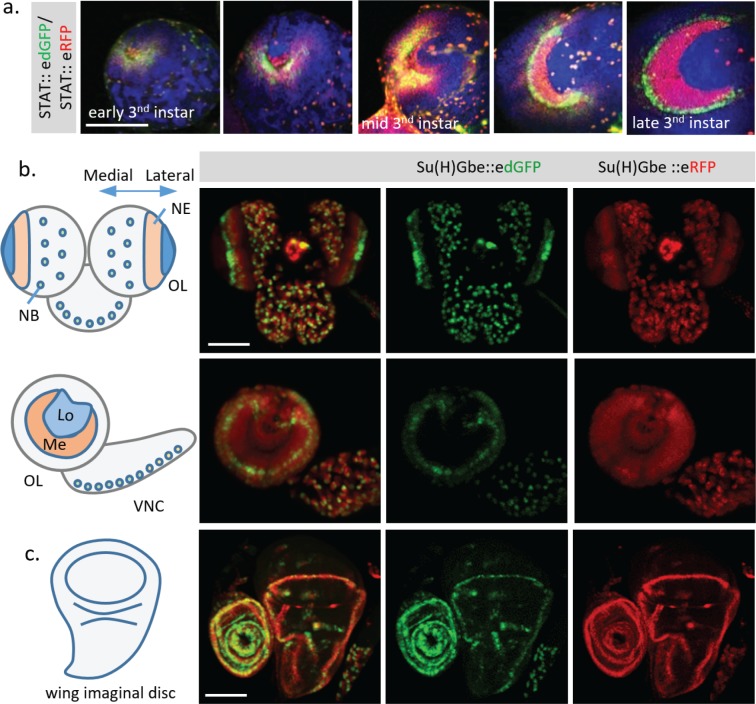
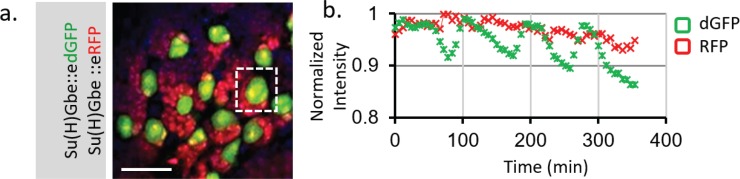
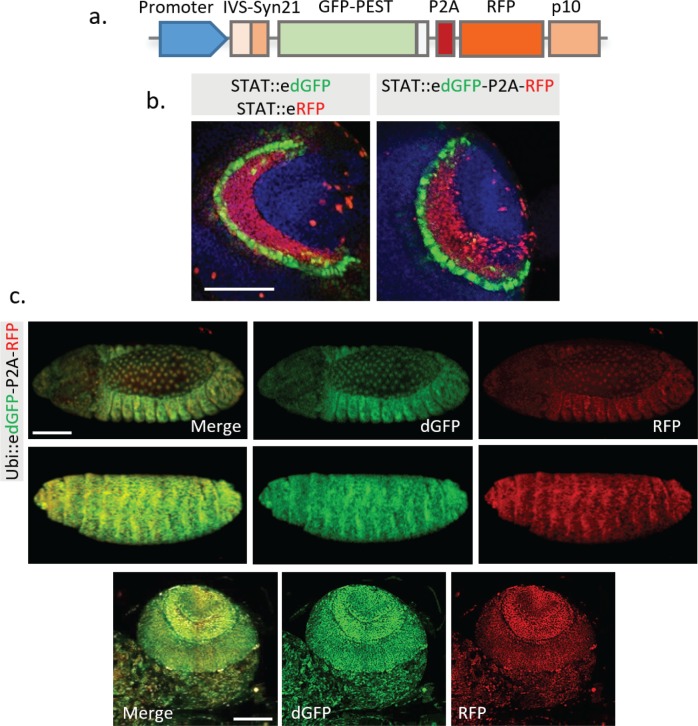
Figure 4. Dynamics of STAT and Notch activity revealed in fixed tissues.
(a) The destabilized GFP (dGFP) combined with stable RFP functions as a fluorescent timer to reveal transcriptional dynamics. The maturation and degradation half-lives of dGFP and RFP are indicated. (b. c) A comparison between the regular GFP reporter with a combination between dGFP and RFP reporter under control of either STAT response elements or Notch response element Su(H)Gbe. Both reporters are visualized in third instar larval brains. NB, neuroblast; GMC, Ganglion mother cells. Scale bar: (b, c) 50 μm.