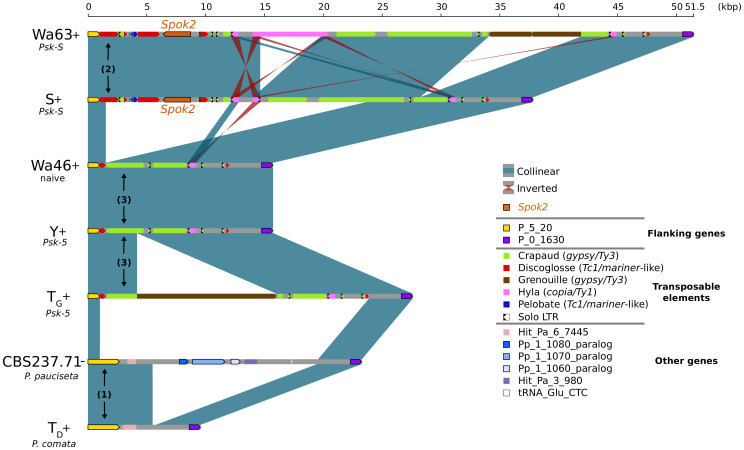
Figure 3. Alignment of the Spok2 locus in selected strains.
The plot displays pairwise comparisons of Spok2 haplotypes with blue segments connecting syntenic regions of homology and red segments showing inversions. The haplotypes are defined by the flanking genes P_5_20 and P_0_1630 located on chromosome 5 of the three sampled species. Every strain has a haplotype of different size, mainly owing to differences in their transposable element (TE) content. Within P. anserina, the TE variation across all sequenced strains occurs downstream of Spok2, as exemplified by strains Wa63 and S. The strains Wa46, Y and TG all lack Spok2 and share break points. See main text for a hypothesis of events (numbered). Notice that P_5_20 stands for the genes Pa_5_20 and PODCO_500020 in the reference annotation of P. anserina and P. comata, respectively, while P_0_1630 stands for Pa_0_1630 and PODCO_001630. As a note, P. pauciseta has a duplication of three genes in tandem from chromosome 1 (Pa_1_1080–60) between the flanking genes. Hit_Pa_X_XXX genes stand for significant BLAST hits to genes of Podan2. TE nomenclature follows Espagne et al. (2008).