Abstract
Purpose:
To develop prospectively accelerated 3D chemical exchange saturation transfer (CEST) imaging using compressed sensing (CS), combined with a saturation scheme based on time-interleaved parallel transmission.
Methods:
A variable density pseudo-random sampling pattern with a centric elliptical k-space ordering was used for CS acceleration in 3D. Retrospective CS studies were performed with CEST phantoms to test the reconstruction scheme. Prospectively CS-accelerated 3D CEST images were acquired in ten healthy volunteers and six brain tumor patients with an acceleration factor (RCS) of 4, and compared to conventional SENSE reconstructed images. Amide proton transfer weighted (APTw) signals under varied RF saturation powers were compared with varied acceleration factors.
Results:
The APTw signals obtained from the CS with RCS = 4 were well preserved as compared with the reference image (SENSE R=2) both in retrospective phantom and prospective healthy volunteer studies. In the patient study, the APTw signals were significantly higher in the tumor region (Gd-enhancing tumor core) than in the normal tissue (p < 0.001). There was no significant APTw difference between the CS-accelerated images and reference image. The scan time of CS-accelerated 3D APTw imaging was dramatically reduced to 2:10 min (in-plane spatial resolution of 1.8 × 1.8mm2; 15 slices with 4 mm slice thickness) as compared with SENSE (4:07 min).
Conclusion:
CS acceleration was successfully extended to 3D CEST imaging without compromising CEST image quality and quantification. The CS-based CEST imaging can easily be integrated into clinical protocols and would be beneficial for a wide range of applications.
Keywords: CEST, APT, compressed sensing, parallel RF transmission, brain tumor
1. INTRODUCTION
Chemical exchange saturation transfer (CEST) imaging is an important molecular MRI method that can generate contrast on free bulk water protons in tissue (1–4) that originates from and reflects labile protons in low concentration (mM range) solute molecules such as metabolites. Amide proton transfer (APT) imaging, a variant of the CEST-based molecular MRI technique, is based on the chemical exchange between free bulk water protons and the amide protons (-NH) of endogenous mobile proteins and peptides in tissue (4–9). An abundance of pre-clinical and clinical data suggests that APT-weighted (APTw) MRI enables the noninvasive identification and differentiation of brain tumors from peritumoral edema or normal tissue, high-grade from low-grade tumors, or radiation necrosis from tumor recurrence (10–18). Notably, the technique has the potential to expand the range of molecular MRI techniques to the endogenous mobile protein and peptide level and has recently become an FDA-approved sequence on 3T MRI systems for clinical use with brain cancers (19).
The early clinical applications of CEST MRI are very promising, but have been limited mainly because of the long scan time required. CEST effects are often assessed using a so-called Z-spectrum, generated by plotting the normalized water signal intensity as a function of RF saturation frequency offsets. The measurement of a complete Z-spectrum based using standard imaging schemes is time-consuming because a large number of RF saturation frequency offsets are inevitably acquired. Similarly, clinical CEST imaging acquisition is also slow because multiple RF saturation frequency offsets are needed for B0 inhomogeneity corrections (20) or CEST quantification (21–25), limiting clinical translation despite of the many clinical benefits. A faster CEST scan would not only help to improve patient comfort and compliance, but also could be used to improve image quality, e.g. by reducing the influence of motion on different saturation frequency offsets. Furthermore, higher spatial resolution imaging would be valuable for assessing tumor heterogeneity more precisely. A fast imaging strategy is thus essential for translating CEST-MRI technology to daily clinical practice. Previous studies demonstrated the feasibility of accelerated CEST MRI using a combined compressed sensing (CS) and sensitivity encoding (SENSE) technique or a learning dictionary-based CS technique in healthy volunteers and/or tumor patients (26–28). However, they were limited by a single-slice acquisition, and a retrospective random sampling scheme.
In CEST MRI, particularly APTw imaging (a relatively slow exchange process on the order of tens to hundreds of Hz), RF saturation time is usually a few seconds in animal studies to obtain steady-state CEST contrast (29–31); however, it is typically limited to <1 sec on human scanners due to hardware constraints, particularly the RF amplifier duty cycle and maximum pulse duration (32–34). To overcome this issue, a time-interleaved parallel transmission (pTX) technique was designed, which supports RF saturation at 100% RF duty-cycle over multiple seconds for highly sensitive detection of APT effects on clinical scanners (35–37). Although a major portion of the CEST imaging time is deployed in the RF saturation period, a significant scan time reduction can be achieved in a 3D imaging sequence using an undersampled k-space acquisition. In this study, a CS-based accelerated CEST imaging technique was developed and tested using a retrospective study in CEST phantoms and prospective studies in healthy volunteers and tumor patients at a 3T human scanner.
2. THEORY
The undersampled k-space data acquired from each coil channel can be reconstructed by solving the following optimization problem, where we minimize the linear combination of a data fidelity term, a total variation term, and sparse wavelet regularization (38):
| [1] |
where Fu is the undersampled Fourier transform, x is the image to be reconstructed, b is the undersampled k-space data, and ψ is a sparsifying wavelet transform. The total variation (TV) is defined as, where in the total variation are the forward finite difference operators along x, y, and z coordinates. The L1-norm defined as the sum of the complex modulus can be estimated by, where μ is positive and close to zero. In the formulation in Eq. [1], the image can be sparse in the transform domains (and), whose tradeoff between the residual norm and regularity is controlled by the constants, α and λ.
3. METHODS
3.1. Phantom Experiment
Tissue-like phantoms were prepared in a 50 mL tubes and placed in a large plastic container that was filled with agarose to minimize field inhomogeneity. Ammonium chloride (NH4Cl, Sigma Aldrich, St Louis, MO, USA) was added in 50 mL tubes by dissolving 500 mM or 1 M ammonium chloride in 40 mL PBS (pH 7.0) and titrated to pH values of 5.5, 6, and 6.5 (39). Furthermore, 1% agarose (Sigma Aldrich, St Louis, MO, USA) was added to tubes, which was heated briefly using a microwave to dissolve agarose. The phantoms were left to solidify at room temperature before MRI scans. For CEST imaging, conventional Z-spectra (normalized water saturation spectra in which Z(Δω)= Ssat(Δω)/S0) were acquired with 49 frequency offsets Δω between −6 ppm and 6 ppm from the water resonance at intervals of 0.25 ppm. RF saturation powers of 1.5, 2.5, and 3.5 μT and a saturation duration of 0.8 sec (a series of four block pulses, 0.2 sec duration each and 100% duty-cycle) were used. Note that parallel RF transmission was not used in the phantom study. CS was retrospectively simulated on the phantom data to test the CS technique prior to the performance of prospective CS acquisitions. The fully sampled k-spaces were retrospectively undersampled with a variable density pseudo-random sampling pattern by a factor (RCS) of 4 and 6. All studies were conducted on a 3 T Achieva dStream system (Philips Healthcare, Best, The Netherlands) with a 32-channel head coil, and all image processing and reconstruction algorithms were implemented in MATLAB R2018 (The MathWorks, Inc., Natick, MA) on a 64-bit Window operation system, with a 16-core 3.4 GHz AMD processor and 64 GB of memory.
3.2. In-vivo MRI Experiments
Ten healthy volunteers (seven males, three females; median age, 26 years old) and six tumor patients (five males, one female; median age, 43 years old) with pathology-confirmed high-grade gliomas (5 recurrent glioblastoma, 1 anaplastic oligodendroglioma) were recruited for this study. The study was approved by the Johns Hopkins Institutional Review Board. Before involvement in this study, written, informed consent was obtained from all participants. A 3D turbo spin echo (TSE)-based CEST sequence was implemented and combined with two parallel RF transmission channels (via a body coil) driven by the RF amplifiers in a time-interleaved fashion. By combining two amplifiers, each operating at 50% duty-cycle, an RF saturation pulse at 100% duty-cycle is achieved for durations up to 5 sec, while the full power of a single RF amplifier is available for the maximum B1 amplitude (37). The CEST images were acquired with 9 frequency offsets (S0, ±3, −3.5, +3.5, +3.5, +3.5, and ±4 ppm), RF saturation powers of 1.5 and 2 μT for healthy volunteers and 2 μT only for patients, and an RF saturation duration of 2 sec. The saturated image at 3.5 ppm was repeated three times with different echo shifts (−0.4, 0, +0.4 msec) for Dixon-type B0 mapping (40). Other imaging parameters were TR/TE = 6.5 sec/6 msec, FOV = 212 × 186 × 60 mm3, spatial resolution = 1.8 × 1.8 × 4 mm3, non-slice-selective 120° or 40° refocusing pulses, turbo factor = 174 or 104, slice oversampling factor = 1.4. A spectral pre-saturation with inversion recovery (SPIR)-based fat suppression was applied after RF saturation preparation. Uniformly undersampled reference (SENSE only, RSENSE = 2 in A-P) and prospectively 4-fold undersampled CS images were acquired. A variable density pseudo-random sampling pattern with a centric elliptical k-space ordering was used for CS acceleration in the two phase encoding directions of the 3D acquisition (Fig. 1A). The identical subsampling pattern was applied along the direction of the saturation frequency offset. Several standard sequences were performed for each patient, but accelerated 8-fold by SENSE (2-fold) and CS (4-fold), including 3D T2-weighted TSE (TR/TE = 3 sec/271 msec; FOV = 212 × 186 × 132mm3; spatial resolution = 1 × 1 × 1mm3; slices =132; scan time = 1:37 min), 3D FLAIR TSE (TR/TE = 4.8 sec/271 msec; FOV = 212 × 186 × 132mm3; spatial resolution = 1 × 1 × 1mm3; slices =132; scan time = 50 sec), 3D T1-weighted TSE (TR/TE = 600 msec/28 msec; FOV = 212 × 186 × 132mm3; spatial resolution = 1 × 1 × 1mm3; slices =132; scan time = 2:10min), and gadolinium enhanced T1-weighted imaging for brain tumor patients using the same parameters. The total scan time of the tumor patient was under 10 min.
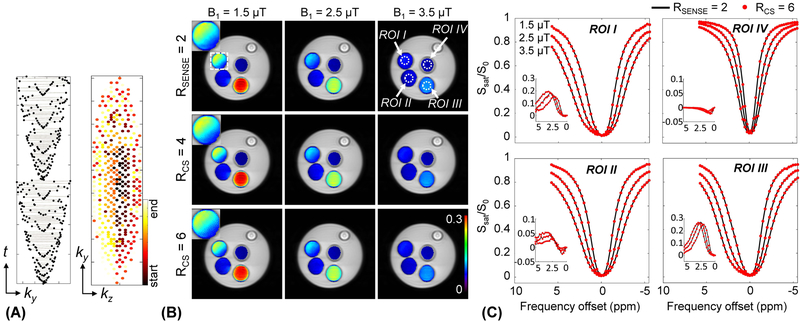
Figure 1.
(A) A variable density k-space undersampling scheme with the centric elliptical k-space ordering for 3D acquisition. The central region is densely sampled and the outer region is progressively undersampled by the acceleration factor (e.g., RCS = 4). The colormap represents echo ordering acquisition (start of acquisition and end of acquisition). Comparison of (B) the MTRasym(2.5ppm) maps and (C) Z-spectra and MTR asymmetry spectra (insets) reconstructed with SENSE with RSENSE = 2 and CS with RCS = 4 (maps) and 6 (maps and spectra). Phantom contained NH4Cl and 1% agarose (ROI I: pH 4.5, 0.5 M NH4Cl + 1% agarose + PBS, ROI II: pH 5.0, 0.5 M NH4Cl + 1% agarose + PBS, ROI III: pH 4.6, 1 M NH4Cl + 1% agarose + PBS, ROI IV: pH 7.0, 1% agarose + PBS).
3.3. MR Image Reconstruction and Analysis
Sensitivity maps for the multi-channel coil were acquired from a separate calibration scan. Iterative reconstruction was used to solve an inverse problem with a sparsity constraint. Image reconstruction was performed using the Daubechies-4 wavelet as the sparse transform and TV penalty to reduce the background noise. The weights for TV and L1 minimization were 0.001 and 0.04, respectively. The weights were chosen to be in a range that minimized the error between the accelerated images and the reference images (RSENSE = 2). Detailed reconstruction methods can be found in a previous publication (26). All CS-reconstructed CEST image volumes were registered to the mean saturated volume (+3.5ppm) and corrected for B0 inhomogeneity effects (41). Lastly, APTw images were generated using magnetization transfer ratio asymmetry analysis, MTRasym(3.5ppm) (5). In terms of quantitative comparison, the normalized root mean squared error (nRMSE) and mean structural similarity index measure (SSIM) were used to estimate the fidelity of accelerated APTw images against the reference. The nRMSE is defined as ǁa - bǁ2/(bmax - bmin), where a is a reference data, b is a test data, and ǁ ǁ2 denotes the L2-norm. The SSIM is defined as (2μa μa+C1)(σab+C2)/(μa2+ μb2+C1)(σa2+ σb2+C2), where μ and σ2 are the mean intensity and variance, respectively. Constants C are included to avoid instabilities when the denominator is very close to zero. A region of interest (ROI) analysis was performed to compare APTw signal intensities between the normal-appearing white matter and glioma. The two ROIs were carefully drawn on the Gd-T1w images which were registered to the APTw images. Tumor ROI definition was guided by the Gd-enhancing regions of the glioma. In addition, ROI-based linear correlation and Bland-Altman analyses were used to compare the intensities from accelerated APTw images to those from reference SENSE-reconstructed APTw images.
4. RESULTS
4.1. Phantom results
Retrospectively sampled data of the ammonium chloride phantom was successfully reconstructed for RCS = 4 and RCS = 6. Figure 1 shows (B) MTRasym(2.5ppm) maps and (C) Z-spectra reconstructed using CS as well as SENSE reconstructions (RSENSE = 2) as a reference. For the Z-spectra, only RCS = 6 is depicted. The CEST signal intensities derived by the three reconstructions at different accelerations were in close agreement for different RF saturation powers and ROIs with varied pH and NH4Cl concentration. The average nRMSE values of the MTRasym(2.5ppm) maps with three RF saturation powers were 0.0048 and 0.0085 for RCS = 4 and RCS = 6, respectively. The average SSIM values of all Z-spectra were as high as 0.99 at a sampling rate of 6. Even though the mean values of the MTRasym(2.5ppm) signal intensities obtained from ROI-based analyses were statistically equivalent (p < 0.05), it can be visually observed that the MTRasym(2.5ppm) maps with RCS = 6 suffer from some minor streaking artifacts which slightly degrade the image quality (magnified images shown in the white dashed square in Fig. 1B). We therefore used RCS = 4 in the human scanning.
4.2. Healthy volunteer results
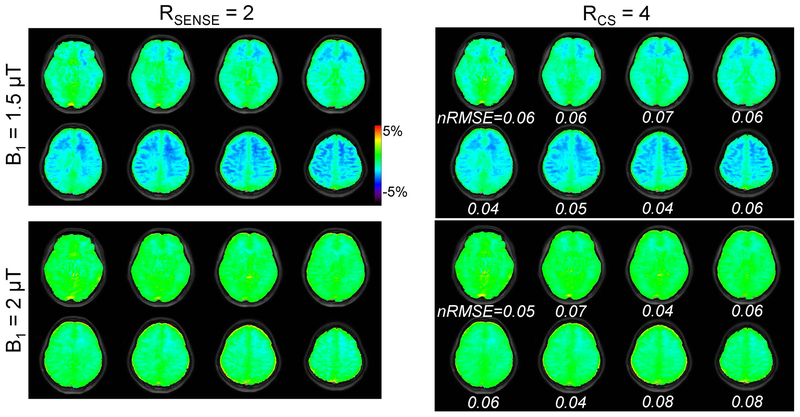
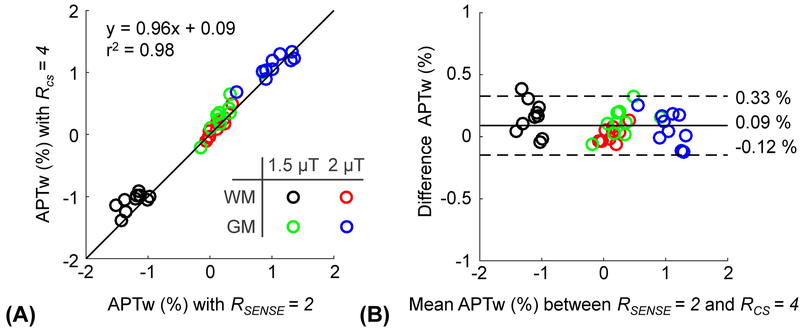
Figure 2 shows the 3D APTw images obtained from conventional SENSE with 2-fold and CS with 4-fold acceleration. The prospective CS acceleration achieved image quality comparable to the conventional SENSE-reconstructed images, but with a scan time reduction of 50% (2:10 min vs. 4:07 min). Interestingly, the APTw signal contrast between gray matter and white matter decreased with increased RF saturation power, which is due to direct water saturation, MTC, water T1 relaxation effects. However, the APTw images obtained from the two RF saturation powers had similar nRMSE values (also see Supporting Information Figure S1) and the RF saturation power was irrelevant for the performance of CS reconstruction. The average nRMSE value for all volunteers was 0.06. The APTw signal intensities of the gray and white matter regions with RF saturation powers of 1.5 and 2 μT were measured for both CS-acceleration and SENSE-acceleration and are compared in Fig. 3. The correlation between the two acceleration methods was high with r2 of 0.98 and a slope close to one (Fig. 3A). The Bland-Altman analysis (Fig. 3B) resulted in significantly but small bias with an overall mean difference of 0.09% (p < 0.05).
Figure 2.
A comparison of APTw images from a healthy volunteer reconstructed with SENSE and CS using prospective acceleration factors of 2 and 4, respectively. Only odd numbered slices are shown. The APTw images were obtained from RF saturation powers of 1.5 and 2 μT, and an RF saturation duration of 2 sec.
Figure 3.
(A) Correlation and (B) Bland-Altman plots of the APTw signal at RF saturation powers of 1.5 and 2 μT, reconstructed from the reference standard (SENSE = 2) and CS factor 4 for ten healthy volunteers. In the Bland-Altman plot, the mean bias is indicated by the solid black line and the 97% limits of agreement are indicated by dotted black lines.
4.3. Patient results
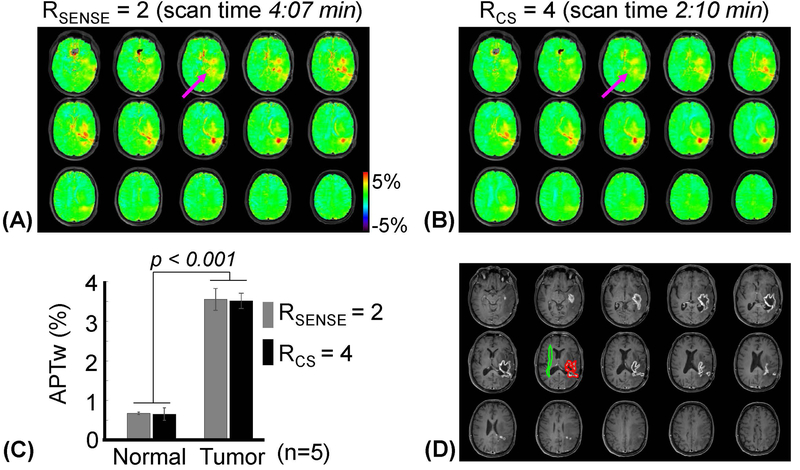
APTw images of a representative patient with recurrent glioblastoma obtained with an RF saturation power of 2 μT and RF saturation time of 2 sec are shown in Figs. 4A and 4B. There was no major visual difference (mean nRMSE = 0.07 and SSIM = 0.93) in the image quality between 2-fold SENSE and 4-fold CS scans (Fig. 4C and also see Supporting Information Figure S2), even though there were mild blurring effects on the CS-reconstructed APTw images (magenta arrows in Figs. 4A and 4B). The two accelerated APTw images had excellent agreement in identifying active tumor enhancements, showing clear APTw hyperintensity in the area of post-Gd enhancement (tumor core) as quantified in Fig. 4D. From five GBM patients, the mean APTw signal intensities for the normal appearing white matter measured from reference and CS-accelerated images were 0.67 ± 0.03 % (95% confidence interval = 0.63% ~ 0.72%) and 0.66 ± 0.14 % (95% confidence interval = 0.40% ~ 0.88%), respectively. The mean APTw signal intensities for the tumor measured from reference and CS-accelerated images were 3.55 ± 0.27 % (95% confidence interval = 3.11% ~ 3.99%) and 3.51 ± 0.19 % (95% confidence interval = 3.20% ~ 3.82%), respectively. The APTw signal intensity of the tumor was significantly higher than that of normal tissue (p < 0.001 for both accelerations, n = 5). Figure 5 shows 3D conventional MR and APTw images for a patient with an anaplastic oligodendroglioma. An intermediate APTw signal intensity was observed in the same tumor area where Gd-T1w image showed faint enhancement.
Figure 4.
APTw images obtained for a patient with a glioblastoma using a RF saturation power of 2 μT and saturation duration of 2 sec, reconstructed by (A) SENSE (RSENSE = 2) and (B) CS (RCS = 4). (C) Average APTw signal intensities with the two acceleration schemes from normal tissue and glioma tissue (from five patients with a glioblastoma). Error bars depict standard deviations. (D) Post-contrast T1w images.
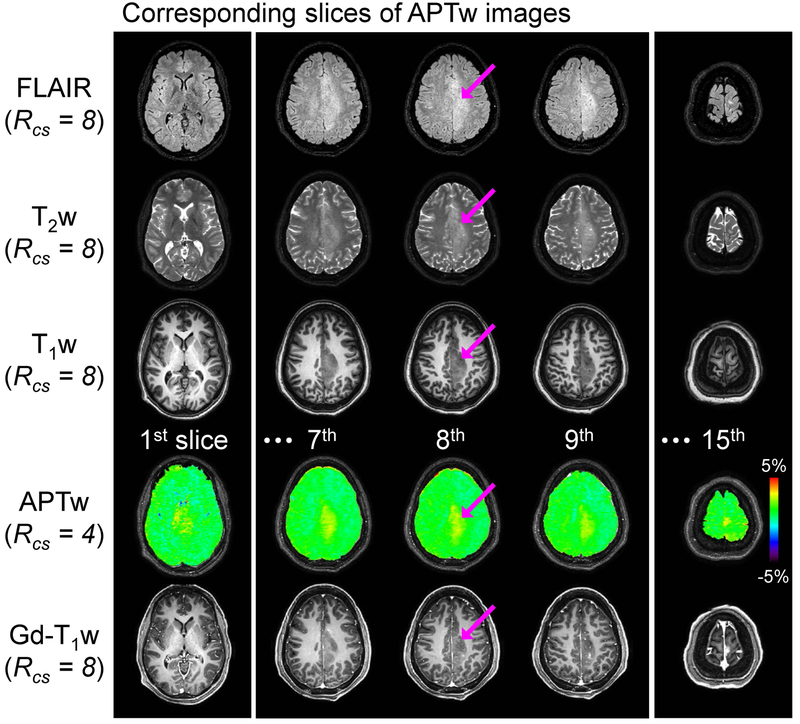
Figure 5.
Conventional MR images and APTw images for a representative patient with an anaplastic oligodendroglioma. Five out of a total of fifteen APTw image slices are shown together with corresponding FLAIR, T2w, T1w, and Gd-T1w images. The grade III lesion was slightly enhanced on the post-contrast images and also showed an intermediate APTw signal.
5. DISCUSSION
Although the clinical applications for CEST or APT-MRI are very promising, a major limitation for routine clinical use is the time requirement and clinical tolerability to add another scan to an already long clinical protocol. The current 3D APTw MRI protocol (~ 15 slices for 6 mm slice thickness) is performed within a clinically acceptable scan time (~ 4min). Whole-brain 3D APTw MRI with higher spatial resolution is particularly important for depicting intratumoral heterogeneity for patients with very large tumor regions, which will take an even longer scan time. Therefore, the development of fast CEST image techniques is a high priority to improve APT-MRI for daily clinical practice (26–28, 42–45). Unlike traditional strategies to accelerate MR image acquisition, CS techniques can reduce scan time by the combination of data sensing, compression, and processing in one operation (38). Herein, the accelerated CEST imaging technique using CS was developed, evaluated using the phantoms, healthy volunteers with varied RF saturation powers, and applied to tumor patients. In-vivo human studies, the 4-fold acceleration was achieved without compromising the quality of APTw images. However, we found that areas around CSF showed larger nRMSE values (see Supporting Information Figure S2) probably because of possible CSF flow-related artifacts, partial volume effects, or suboptimal image registration results. In the healthy volunteer studies, the APTw signal contrast between gray and white matter decreased with RF saturation power. The most likely explanation of this is differences in amide, nuclear Overhauser enhancement (NOE), and semisolid macromolecular proton concentrations, and water T1 relaxation time between gray and white matters, leading to very different contributions with RF saturation powers (46, 47). However, the RF saturation power of 2 μT was used in the tumor patient studies because the power is ideal for APTw imaging at clinical 3T magnetic field strengths.
Compared with our previous work, several novel technical aspects were presented in the current study. To the best of our knowledge, this is the first time that k-space measurements were prospectively undersampled and reconstructed using CS reconstruction in CEST imaging. We extended the CS reconstruction to prospectively undersampled 3D APTw data and successfully applied this to brain tumor patient imaging. The scan time of 3D APTw imaging was 2:10 min for 15 slices (4 mm thickness) with an in-plane spatial resolution of 1.8 × 1.8mm2 and the CS reconstruction was performed in less than 2 min. Second, the use of the time-interleaved parallel transmission in the RF saturation scheme allowed us to maximize APT contrast using 100% duty-cycle saturation over 2 seconds (see Supporting Information Figure S3). With a pTX-based pseudo continuous-wave (CW) RF irradiation on 3T clinical scanners, a simple analytical solution of the proton exchange model can be derived from full Bloch-McConnell equations. Furthermore, the Dixon approach provides robust B0 inhomogeneity correction and can be achieved during the APT acquisition and no extra scans are needed for B0 correction.
Our CS APT-MRI protocol enabled acceleration by a factor of RCS = 4, or potentially to improve image quality (higher spatial resolution or spatial coverage) while maintaining the acceleration rates. However, static, sparsifying, transform-based CS still suffers from several artifacts at high undersampling factors. To accurately reconstruct accelerated images, solution of an optimization problem with multiple mathematical constraints is required. An adaptive transform (learned dictionary from training data) applied in the CS framework may lead to higher image sparsity, and thus, to a potentially higher acceleration factor than the conventional, global, sparsifying transform commonly used in classic CS, even though this processing proves computationally complex and time-consuming (27). For the sake of simplicity, in the current study, the identical subsampling pattern was used along frequency offset domain. However, CS acceleration can be further increased by exploiting the sparsity in the frequency offset dimension with varied sub-sampling patterns for each Z-spectral image, in a similar way to incoherent sampling patterns in parameter dimensions in CS-based MR T1 and T2 parameter mapping (48). Our additional retrospective phantom study showed a significant improvement in reconstruction accuracy when random sampling patterns are applied along the frequency offset direction (see Supporting Information Figure S4 and S5). Recently, deep learning approaches have been used to model image reconstruction problems by training deep neural networks, which take full k-space measurements as inputs and put out reconstructed images, which is a non-iterative process after training occurs, thus achieving faster reconstruction (49–51).
A variable-density sampling pattern with an elliptic-centric 3D k-space ordering was used for accelerated APTw-MRI, which typically yields better quantitative measures in terms of de-noising and artifacts because most of the energy of images is concentrated in the center of k-space. However, slight blurring effects were visible in the accelerated APTw images of the healthy volunteer and the tumor patient due to the high undersampling of the outer lines of k-space and the denoising properties of CS reconstruction (38). Therefore, further studies are still needed to find an optimal sampling pattern of the k-space.
6. CONCLUSIONS
The CS-based CEST MRI technique was successfully extended from 2D to 3D, achieving a prospective acceleration factor of 4, and offered a significant scan time reduction. Accelerated CEST MRI with CS, combined with a parallel transmission-based saturation scheme could help to facilitate the clinical translation of CEST MRI into a routine diagnostic tool to monitor tumor response and progression of therapy, as well as for application to other diseases, such as hyperacute stroke and other neurological disorders.
7. ACKNOWLEGEMENTS
The authors thank Dr. Zheng Han for the phantom preparation. This work was supported in part by grants from the National Institutes of Health (R21CA227783, R01CA166171, R01CA228188, R01EB015032, and P41EB015909) and the Rad BriteStar award from the Department of Radiology Johns Hopkins University School of Medicine.
Supplementary Material
Supporting Information Figure S1. A comparison of 3D APTw images (15 slices) from a healthy volunteer reconstructed with SENSE and CS using prospective acceleration factors of 2 and 4, respectively.
Supporting Information Figure S2. APTw images obtained for a patient with a glioblastoma using a RF saturation power of 2 μT and saturation duration of 2 sec, reconstructed by (A) SENSE (RSENSE = 2) and (B) CS (RCS = 4). (C) The corresponding error images (= |SENSE - CS|), nRMSE and SSIM values, from comparing the two reconstructions.
Supporting Information Figure S3. APTw images obtained for a patient with a glioblastoma using a RF saturation power of 2 μT and saturation durations of 0.8, 2, and 2.4 sec at 100% RF duty-cycle.
Supporting Information Figure S4. MTRasym(2.5ppm) maps reconstructed by SENSE with RSENSE = 2 (top row) and CS with RCS = 8 (middle row: identical sampling pattern applied along a frequency offset direction and bottom row: random sampling pattern applied along a frequency offset direction).
Supporting Information Figure S5. Correlation and Bland-Altman plots of the MTRasym(2.5ppm) signal at RF saturation powers of 1.5 μT, reconstructed from the reference standard (SENSE = 2) and CS factor 8 (top row: random sampling pattern applied along a frequency offset direction and bottom row: identical sampling pattern applied along a frequency offset direction). In the Bland-Altman plot, the mean bias is indicated by the solid black line and the 97% limits of agreement are indicated by dotted black lines.
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article.
REFERENCE
- 1.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J Magn Reson 2000;143:79–87. [DOI] [PubMed] [Google Scholar]
- 2.Sherry AD, Woods M. Chemical exchange saturation transfer contrast agents for magnetic resonance imaging. Annu Rev Biomed Eng 2008;10:391–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones KM, Pollard AC, Pagel MD. Clinical applications of chemical exchange saturation transfer (CEST) MRI. J Magn Reson Imaging 2018;47:11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou J, Heo HY, Knutsson L, van Zijl PCM, Jiang S. APT-weighted MRI: Techniques, current neuro applications, and challenging issues. J Magn Reson Imaging 2019:DOI: 10.1002/jmri.26645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J, Payen J, Wilson DA, Traystman RJ, van Zijl PCM. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nature Med 2003;9:1085–1090. [DOI] [PubMed] [Google Scholar]
- 6.Sun PZ, Zhou J, Sun W, Huang J, van Zijl PCM. Detection of the ischemic penumbra using pH-weighted MRI. J Cereb Blood Flow Metab 2007;27:1129–1136. [DOI] [PubMed] [Google Scholar]
- 7.By S, Barry RL, Smith AK, Lyttle BD, Box BA, Bagnato FR, Pawate S, Smith SA. Amide proton transfer CEST of the cervical spinal cord in multiple sclerosis patients at 3T. Magn Reson Med 2018;79:806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan K, Fu Z, Yang C, Zhang K, Jiang S, Lee DH, Heo HY, Zhang Y, Cole RN, Van Eyk JE, Zhou J. Assessing amide proton transfer (APT) MRI contrast origins in 9L gliosarcoma in the rat brain using proteomic analysis. Mol Imaging Biol 2015;17:479–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang S, Yu H, Wang X, Lu S, Li Y, Feng L, Zhang Y, Heo HY, Lee DH, Zhou J, Wen Z. Molecular MRI differentiation between primary central nervous system lymphomas and high-grade gliomas using endogenous protein-based amide proton transfer MR imaging at 3 Tesla. Eur Radiol 2016;26:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J, Zhu H, Lim M, Blair L, Quinones-Hinojosa A, Messina AA, Eberhart CG, Pomper MG, Laterra J, Barker PB, van Zijl PCM, Blakeley JO. Three-dimensional amide proton transfer MR imaging of gliomas: Initial experience and comparison with gadolinium enhancement. J Magn Reson Imaging 2013;38:1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heo HY, Zhang Y, Jiang S, Lee DH, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging with extrapolated semisolid magnetization transfer reference (EMR) signals: II. Comparison of three EMR models and application to human brain glioma at 3 Tesla. Magn Reson Med 2016;75:1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang S, Eberhart CG, Zhang Y, Heo H-Y, Wen Z, Blair L, Qin H, Lim M, Quinones-Hinojosa A, Weingart JD, Barker PB, Pomper MG, Laterra J, van Zijl PCM, Blakeley JO, Zhou J. Amide proton transfer-weighted MR image-guided stereotactic biopsy in patients with newly diagnosed gliomas. Eur J Cancer 2017;83:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Tryggestad E, Wen Z, Lal B, Zhou T, Grossman R, Wang S, Yan K, Fu D-X, Ford E, Tyler B, Blakeley J, Laterra J, van Zijl PCM. Differentiation between glioma and radiation necrosis using molecular magnetic resonance imaging of endogenous proteins and peptides. Nature Med 2011;17:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong X, Liu L, Wang M, Ding K, Fan Y, Ma B, Lal B, Tyler B, Mangraviti A, Wang S, Wong J, Laterra J, Zhou J. Quantitative multiparametric MRI assessment of glioma response to radiotherapy in a rat model. Neuro-Oncology 2014;16:856–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma B, Blakeley JO, Hong X, Zhang H, Jiang S, Blair L, Zhang Y, Heo HY, Zhang M, van Zijl PC, Zhou J. Applying amide proton transfer-weighted MRI to distinguish pseudoprogression from true progression in malignant gliomas. J Magn Reson Imaging 2016;44:456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JE, Kim HS, Park KJ, Kim SJ, Kim JH, Smith SA. Pre- and Posttreatment Glioma: Comparison of Amide Proton Transfer Imaging with MR Spectroscopy for Biomarkers of Tumor Proliferation. Radiology 2016;278:514–523. [DOI] [PubMed] [Google Scholar]
- 17.Jiang S, Eberhart CG, Lim M, Heo HY, Zhang Y, Blair L, Wen Z, Holdhoff M, Lin D, Huang P, Qin H, Quinones-Hinojosa A, Weingart JD, Barker PB, Pomper MG, Laterra J, van Zijl PCM, Blakeley JO, Zhou J. Identifying Recurrent Malignant Glioma after Treatment Using Amide Proton Transfer-Weighted MR Imaging: A Validation Study with Image-Guided Stereotactic Biopsy. Clin Cancer Res 2019;25:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Togao O, Yoshiura T, Keupp J, Hiwatashi A, Yamashita K, Kikuchi K, Suzuki Y, Suzuki SO, Iwaki T, Hata N, Mizoguchi M, Yoshimoto K, Sagiyama K, Takahashi M, Honda H. Amide proton transfer imaging of adult diffuse gliomas: correlation with histopathological grades. Neuro Oncol 2014;16:441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.U.S. Food and Drug Administration, https://www.accessdata.fda.gov/cdrh_docs/pdf17/K172920.pdf.
- 20.Zhou J, Blakeley JO, Hua J, Kim M, Laterra J, Pomper MG, van Zijl PCM. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med 2008;60:842–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chappell MA, Donahue MJ, Tee YK, Khrapitchev AA, Sibson NR, Jezzard P, Payne SJ. Quantitative Bayesian model-based analysis of amide proton transfer MRI. Magn Reson Med 2013;70:556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai K, Singh A, Poptani H, Li W, Yang S, Lu Y, Hariharan H, Zhou XJ, Reddy R. CEST signal at 2ppm (CEST@2ppm) from Z-spectral fitting correlates with creatine distribution in brain tumor. NMR Biomed 2015;28:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heo HY, Zhang Y, Lee DH, Hong X, Zhou J. Quantitative assessment of amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging with extrapolated semi-solid magnetization transfer reference (EMR) signals: Application to a rat glioma model at 4.7 tesla. Magn Reson Med 2016;75:137–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee DH, Heo HY, Zhang K, Zhang Y, Jiang S, Zhao X, Zhou J. Quantitative assessment of the effects of water proton concentration and water T1 changes on amide proton transfer (APT) and nuclear overhauser enhancement (NOE) MRI: The origin of the APT imaging signal in brain tumor. Magn Reson Med 2017;77:855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heo HY, Zhang Y, Burton TM, Jiang S, Zhao Y, van Zijl PCM, Leigh R, Zhou J. Improving the detection sensitivity of pH-weighted amide proton transfer MRI in acute stroke patients using extrapolated semisolid magnetization transfer reference signals. Magn Reson Med 2017;78:871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heo HY, Zhang Y, Lee DH, Jiang S, Zhao X, Zhou J. Accelerating chemical exchange saturation transfer (CEST) MRI by combining compressed sensing and sensitivity encoding techniques. Magn Reson Med 2017;77:779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heo H-Y, Bhave S, Jacob M, Zhou J. Blind Compressed Sensing-based Ultrafast Chemical Exchange Saturation Transfer (CEST) Imaging In Proc 24th Annual Meeting ISMRM, Singapore, 2016. p. 301. [Google Scholar]
- 28.She H, Greer JS, Zhang S, Li B, Keupp J, Madhuranthakam AJ, Dimitrov IE, Lenkinski RE, Vinogradov E. Accelerating chemical exchange saturation transfer MRI with parallel blind compressed sensing. Magn Reson Med 2018:doi: 10.1002/mrm.27400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PCM. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med 2003;50:1120–1126. [DOI] [PubMed] [Google Scholar]
- 30.Zu Z, Janve VA, Xu J, Does MD, Gore JC, Gochberg DF. A new method for detecting exchanging amide protons using chemical exchange rotation transfer. Magn Reson Med 2013;69:637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heo HY, Zhang Y, Jiang S, Zhou J. Influences of experimental parameters on chemical exchange saturation transfer (CEST) metrics of brain tumors using animal models at 4.7T. Magn Reson Med 2018: 10.1002/mrm.27389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu H, Jones CK, van Zijl PCM, Barker PB, Zhou J. Fast 3D chemical exchange saturation transfer (CEST) imaging of the human brain. Magn Reson Med 2010;64:638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt B, Zaiß M, Zhou J, Bachert P. Optimization of pulse train presaturation for CEST imaging in clinical scanners. Magn Reson Med 2011;65:1620–1629. [DOI] [PubMed] [Google Scholar]
- 34.Heo HY, Jones CK, Hua J, Yadav N, Agarwal S, Zhou J, van Zijl PC, Pillai JJ. Whole-brain amide proton transfer (APT) and nuclear overhauser enhancement (NOE) imaging in glioma patients using low-power steady-state pulsed chemical exchange saturation transfer (CEST) imaging at 7T. J Magn Reson Imaging 2016;44:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Togao O, Hiwatashi A, Keupp J, Yamashita K, Kikuchi K, Yoshiura T, Yoneyama M, Kruiskamp MJ, Sagiyama K, Takahashi M, Honda H. Amide Proton Transfer Imaging of Diffuse Gliomas: Effect of Saturation Pulse Length in Parallel Transmission-Based Technique. PLoS One 2016;11:doi: 10.1371/journal.pone.0155925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang S, Seiler S, Wang X, Madhuranthakam AJ, Keupp J, Knippa EE, Lenkinski RE, Vinogradov E. CEST-Dixon for human breast lesion characterization at 3 T: A preliminary study. Magn Reson Med 2018;80:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keupp J, Baltes C, Harvey PR, Brink vd. Parallel RF transmission based MRI technique for highly sensitive detection of amide proton transfer in the human brain In Proc 19th Annual Meeting ISMRM, Montreal, Quebec, 2011. p. 710. [Google Scholar]
- 38.Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007;58:1182–1195. [DOI] [PubMed] [Google Scholar]
- 39.Heo HY, Han Z, Jiang S, Schar M, van Zijl PCM, Zhou J. Quantifying amide proton exchange rate and concentration in chemical exchange saturation transfer imaging of the human brain. Neuroimage 2019;189:202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keupp J, Eggers H. CEST-Dixon MRI for sensitive and accurate measurement of amide proton transfer in humans at 3T In Proc 18th Annual Meeting ISMRM, Stockholm, Sweden, 2010. p. 338. [Google Scholar]
- 41.Zhang Y, Heo HY, Lee DH, Zhao X, Jiang S, Zhang K, Li H, Zhou J. Selecting the reference image for registration of CEST series. J Magn Reson Imaging 2016;43:756–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Heo HY, Jiang S, Lee DH, Bottomley PA, Zhou J. Highly accelerated chemical exchange saturation transfer (CEST) measurements with linear algebraic modeling. Magn Reson Med 2016;76:136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Heo HY, Lee DH, Jiang S, Zhao X, Bottomley PA, Zhou J. Chemical exchange saturation transfer (CEST) imaging with fast variably-accelerated sensitivity encoding (vSENSE). Magn Reson Med 2017;77:2225–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varma G, Lenkinski R, Vinogradov E. Keyhole chemical exchange saturation transfer. Magn Reson Med 2012;68:1228–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaiss M, Ehses P, Scheffler K. Snapshot-CEST: Optimizing spiral-centric-reordered gradient echo acquisition for fast and robust 3D CEST MRI at 9.4 T. NMR Biomed 2018;31:e3879. [DOI] [PubMed] [Google Scholar]
- 46.Heo HY, Lee DH, Zhang Y, Zhao X, Jiang S, Chen M, Zhou J. Insight into the quantitative metrics of chemical exchange saturation transfer (CEST) imaging. Magn Reson Med 2017;77:1853–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zu Z Towards the complex dependence of MTRasym on T1w in amide proton transfer (APT) imaging. NMR Biomed 2018;31:e3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doneva M, Bornert P, Eggers H, Stehning C, Senegas J, Mertins A. Compressed sensing reconstruction for magnetic resonance parameter mapping. Magn Reson Med 2010;64:1114–1120. [DOI] [PubMed] [Google Scholar]
- 49.Hammernik K, Klatzer T, Kobler E, Recht MP, Sodickson DK, Pock T, Knoll F. Learning a variational network for reconstruction of accelerated MRI data. Magnet Reson Med 2018;79:3055–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han Y, Yoo J, Kim HH, Shin HJ, Sung K, Ye JC. Deep learning with domain adaptation for accelerated projection-reconstruction MR. Magn Reson Med 2018;80:1189–1205. [DOI] [PubMed] [Google Scholar]
- 51.Hyun CM, Kim HP, Lee SM, Lee S, Seo JK. Deep learning for undersampled MRI reconstruction. Phys Med Biol 2018;63:135007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1. A comparison of 3D APTw images (15 slices) from a healthy volunteer reconstructed with SENSE and CS using prospective acceleration factors of 2 and 4, respectively.
Supporting Information Figure S2. APTw images obtained for a patient with a glioblastoma using a RF saturation power of 2 μT and saturation duration of 2 sec, reconstructed by (A) SENSE (RSENSE = 2) and (B) CS (RCS = 4). (C) The corresponding error images (= |SENSE - CS|), nRMSE and SSIM values, from comparing the two reconstructions.
Supporting Information Figure S3. APTw images obtained for a patient with a glioblastoma using a RF saturation power of 2 μT and saturation durations of 0.8, 2, and 2.4 sec at 100% RF duty-cycle.
Supporting Information Figure S4. MTRasym(2.5ppm) maps reconstructed by SENSE with RSENSE = 2 (top row) and CS with RCS = 8 (middle row: identical sampling pattern applied along a frequency offset direction and bottom row: random sampling pattern applied along a frequency offset direction).
Supporting Information Figure S5. Correlation and Bland-Altman plots of the MTRasym(2.5ppm) signal at RF saturation powers of 1.5 μT, reconstructed from the reference standard (SENSE = 2) and CS factor 8 (top row: random sampling pattern applied along a frequency offset direction and bottom row: identical sampling pattern applied along a frequency offset direction). In the Bland-Altman plot, the mean bias is indicated by the solid black line and the 97% limits of agreement are indicated by dotted black lines.