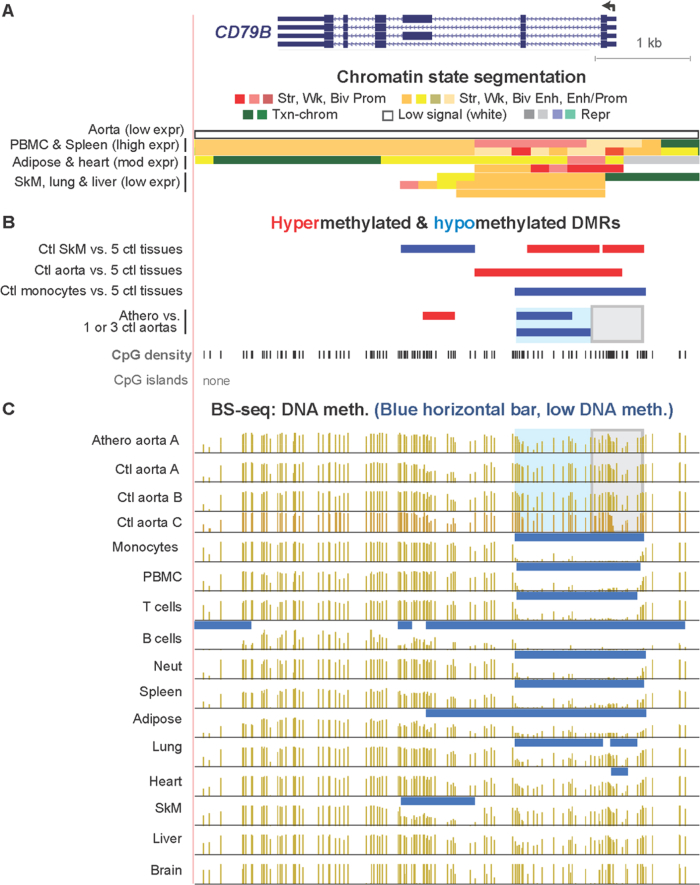
Fig. 1.
CD79B, a B-cell antigen-encoding gene, displays a leukocyte-associated hypomethylated region, which only partly overlaps an atherosclerosis-associated hypomethylated DMR. (A) CD79B is aligned with chromatin state segmentation tracks labeled according to the indicated color code (RefSeq gene isoforms; chr17:62,005,196-62,010,606, hg19). Note the low signal for standard histone modifications throughout this region in aorta (white box). (B) Tissue-specific and atherosclerosis-related DMRs. The density of CpGs is shown; there were no CpG islands in this region according to the UCSC Genome Browser [5]. (C) Bisulfite-seq tracks are plotted as average % methylation at CpGs; blue horizontal bars, regions that display statistically significant hypomethylation relative to the rest of the same genome. Light blue highlighting, the region of atherosclerosis-associated hypomethylation overlapping monocyte- (and leukocyte-) hypomethylation; gray highlighting, the region that did not exhibit an athero hypometh DMR but did show monocyte- (and leukocyte-) hypomethylation. Ctl, control; SkM, skeletal muscle; brain, prefrontal cortex; heart, left ventricle; PBMC, peripheral mononuclear blood cells; neut, neutrophils; expr, expression; str, strong; wk, weak; biv, bivalent; prom, promoter; enh, enhancer; repr, repressed; Txn-chrom, chromatin with a type of histone modification seen in many actively transcribed chromatin regions (histone H3 lysine-36 trimethylation).