Figure 3.
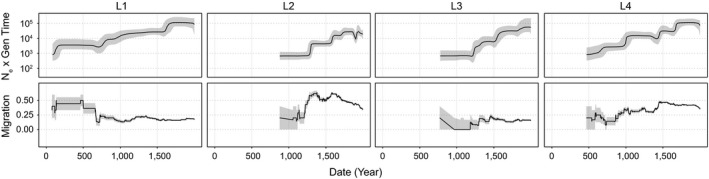
Patterns of effective population size and migration through time of Mycobacterium tuberculosis lineages 1–4. Bayesian skyline plots (top panels) show inferred changes in effective population size (N e) through time deduced from lineage specific analyses. Black lines denote median N e and grey shading the 95% highest posterior density. Estimated migration through time for each lineage is shown in the bottom panels (see Section 2.15). Grey shading depicts the rates inferred after the addition or subtraction of a single migration event, and demonstrate the uncertainty of rate estimates, particularly from the early history of each lineage where branches are coalescing towards the MRCA (data omitted where less than four branches of the phylogeny are represented). Dates are shown in years and are based on scaling the phylogeny of the Old World collection with a substitution rate of 5 × 10−8 substitutions/site/year (Kay et al., 2015)
