Abstract
Pathogenic de novo variants in the X-linked gene SLC35A2 encoding the major Golgi-localized UDP-galactose transporter required for proper protein and lipid glycosylation cause a rare type of congenital disorder of glycosylation known as SLC35A2-CDG (formerly CDG-IIm). To date, twenty-nine unique de novo variants from thirty-two unrelated individuals have been described in the literature. The majority of affected individuals are primarily characterized by varying degrees of neurological impairments with or without skeletal abnormalities. Surprisingly, most affected individuals do not show abnormalities in serum transferrin N-glycosylation, a common biomarker for most types of CDG. Here we present data characterizing 30 individuals and add 26 new variants, the single largest study involving SLC35A2-CDG. The great majority of these individuals had normal transferrin glycosylation. In addition, expanding the molecular and clinical spectrum of this rare disorder, we developed a robust and reliable biochemical assay to assess SLC35A2-dependent UDP-galactose transport activity in primary fibroblasts. Finally, we show that transport activity is directly correlated to the ratio of wild-type to mutant alleles in fibroblasts from affected individuals.
Keywords: Congenital Disorders of Glycosylation (CDG), nucleotide sugar transporter, UDP-galactose, glycoside
INTRODUCTION
Congenital disorders of glycosylation (CDG) are an expanding group of metabolic diseases that primarily present with neurological involvement but can affect all organ systems depending on the specific CDG type (Freeze, Eklund, Ng & Patterson, 2012; Freeze, Eklund, Ng & Patterson, 2015). The majority of these defects can be initially identified by carbohydrate deficient transferrin (CDT) analysis, which examines the glycosylation status of the abundant serum protein, transferrin (Tf) (Lacey, Bergen, Magera, Naylor & O’Brien, 2001; Freeze, 2006). While this is a reliable screening tool for identifying individuals with CDG, it cannot indicate the gene responsible for that disorder. Thus far, the vast majority of CDG types are rare autosomal recessive or X-linked disorders (Ferreira et al., 2018), but as more affected individuals have undergone next generation sequencing (NGS), reports of pathogenic de novo variants are becoming more common (Ng & Freeze, 2018).
SLC35A2 (MIM# 314375) is an X-linked gene, encoding the primary UDP-galactose transporter in humans (Hara et al., 1993; Ishida, Miura, Yoshioka & Kawakita, 1996; Miura et al., 1996). It belongs to the SLC35A family of nucleotide sugar transporters, which also includes the characterized transporters, SLC35A1 (CMP-sialic acid) and SLC35A3 (UDP-N-acetylglucosamine) (Deutscher, Nuwayhid, Stanley, Briles & Hirschberg, 1984; Guillen, Abeijon & Hirschberg, 1998), as well as two putative nucleotide sugar carriers SLC35A4 and SLC35A5 with unknown substrate specificity.
Using mammalian cell-based models, it has been demonstrated that inactivation of SLC35A2 protein abolishes transport of UDP-galactose into the Golgi apparatus, resulting in the synthesis of truncated glycans lacking galactose (Brandli, Hansson, Rodriguez-Boulan, Simons, 1998; Ishida et al., 1999; Oelmann, Stanley & Gerardy-Schahn, 2001). These include N- and O-linked glycans, glycosaminoglycans (GAG) and glycosphingolipids (GSL) (Brockhausen & Stanley, 2015; Lindahl, Couchman, Kimata & Esko, 2015; Schnaar & Kinoshita, 2015; Stanley, Taniguchi & Aebi, 2015). In 2013, whole exome sequencing (WES) identified de novo SLC35A2 variants in the first three individuals with SLC35A2-CDG (MIM# 300896) (Ng et al., 2013). The biochemical analysis of primary fibroblasts from affected individuals supported pathogenicity of these variants. (Ng et al., 2013).
To date, molecular and clinical information on 32 individuals with de novo variants in SLC35A2 have been reported with most exhibiting neurological symptoms, especially epilepsy, developmental delay and intellectual disability (Ng et al., 2013; Kodera et al., 2013; EuroEPINOMICS-RES Consortium, Epilepsy Phenome/Genome Project & Epi4K Consortium, 2014; Dorre et al., 2015; Bosch et al., 2016; Lelieveld et al., 2016; Kimizu et al., 2017; Bruneel et al., 2018; Westenfield et al., 2018; Winawer et al., 2018; Yates et al., 2018; Sim et al., 2018). Recently, WES analysis of brain specimens from 56 individuals identified five subjects harboring somatic de novo SLC35A2 variants (Winawer et al., 2018).
SLC35A2 is X-linked and most affected individuals are females. This is an important factor that could account for the variations seen in both the biochemical and clinical phenotypes due to X-chromosome inactivation (XCI). Females are inherently mosaic for X-linked genes because XCI will randomly silence one of their two allele. This random inactivation can ultimately determine the degree to which an allele is expressed (Galupa & Heard, 2018).
In this study, we characterized the molecular, clinical and biochemical data from a cohort of 30 individuals with SLC35A2-CDG and developed a biochemical assay to reliably measure SLC35A2-dependent UDP-galactose transport into the Golgi apparatus of primary fibroblasts.
MATERIALS and METHODS
Subjects and Clinical Information
Families included in our SLC35A2-CDG research study provided written consent under an approved Sanford Burnham Prebys Medical Discovery Institute IRB protocol or an approved IRB through each medical institution. The only inclusion criteria for this study were either the identification of de novo variants within SLC35A2 or the identification of an abnormal carbohydrate deficient transferrin (CDT) test result suggestive of a galactosylation defect. Primary fibroblasts were grown from a skin biopsy that was obtained by the subjects’ physician.
Molecular Analysis of SLC35A2
Variant analysis for SLC35A2 (NM_001042498.2, ENST00000376521.6) was carried out using either direct Sanger sequencing of the five coding exons or in many instances next generation sequencing (i.e. whole exome sequencing, whole genome sequencing, targeted gene panels). PCR primers and conditions are available upon request. Next generation sequencing was performed at multiple institutions, both commercial and academic on different platforms; however, all variants were Sanger confirmed for all probands and available parents. All variants have been submitted to the LOVD database (https://databases.lovd.nl/shared/genes/SLC35A2).
In vitro UDP-Galactose Transport Assay
Chinese Hamster Ovary (CHO), CHO-Lec8 (Stanley, 1981) and primary fibroblasts (Ichikawa, Scott, Losfeld & Freeze, 2014) were cultured as described. CHO-Lec8 bearing a c.275_374del was used in all experiments and are available from American Type Culture Collection (ATCC CRL-1737) (Stanley, 1981; Oelmann, Stanley & Gerardy-Schahn, 2001). Cells were harvested at 70–80% confluence and permeabilized using hypotonic conditions as described (Kim, Miura, Etchison & Freeze, 2001). Initially the glycoside concentration, temperature and time of the assay were optimized using CHO and CHO-Lec8 cells. Thereafter, UDP-galactose transport was always carried out for 30 min at 37°C using 27 nM UDP-[6-3H] galactose (60Ci/mmol - American Radiolabeled Chemicals, Inc Saint Louis, MO) as a donor substrate and 1 mM 4-Methylumbelliferyl β-N-acetylglucosamine (GlcNAcβ−4-MU; Sigma Aldrich, Saint Louis, MO) as an artificial acceptor in DMSO (Sigma Aldrich, Saint Louis, MO), final concentration of 1%. Prior to adding UDP-[6-3H] galactose to the reaction, permeabilized cells were pre-incubated for 5 minutes with GlcNAcβ−4-MU at 37°C to allow penetration of the glycoside. As a negative control, equal volume of DMSO (final concentration 1%) was added instead of the glycoside. After 30 minutes the reaction product, [6-3H] Galβ1,4GlcNAcβ−4-MU was extracted from the cells with 70% ethanol, purified on Sep-Pak 0.2 ml C18 columns (Waters Corporation, Milford, MA) and the radioactive product measured as previously described (Kim, Miura, Etchison & Freeze, 2001). For data statistical analysis, one-way analysis of variance (ANOVA) test with Tukey’s post hoc test was employed. All analyses were performed with GraphPad Prism (GraphPad Software, La Jolla, CA). Statistical significance was assigned to p-value <0.05.
Determination of kinetic parameters were performed in CHO cells under optimized conditions, using increasing concentrations of unlabeled UDP-galactose (Sigma Aldrich, Saint Louis, MO) with 0.25μCi 3H substrate. Michaelis constant and Vmax for UDP-galactose transport were calculated as disintegrations per minute (dpm) and determined using non-linear regression fit in GraphPad Prism.
SLC35A2 cDNA Allele Ratios
A total of 500 ng RNA was isolated from primary fibroblasts using TRIZOL reagent (Thermo Fisher Scientific, Waltham, MA) and used for cDNA synthesis with QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany) according the manufacturer’s instruction. To determine the ratio of wild-type (WT) and mutant SLC35A2 alleles, specific primers were designed to PCR amplify the region where each individual’s mutated transcript was located. Importantly, each variant results in either a gain or loss of the indicated restriction site (Supp. Figure S1) within the PCR product. Restriction digestion was performed for 1 hour at the temperature that is optimal for each enzyme, according to the manufacturer’s instructions (New England Biolabs, Ipswich, MA). As a control, PCR product amplified on cDNA derived from the subjects cells was treated the same way, but restriction enzyme was replaced with an equal volume of water. Both digested and undigested PCR products were separated on 4% agarose gel and visualized with ethidium bromide. ChemiDoc Imaging System (BioRad, Hercules, CA) was used to document the gels. Wild type to mutant allele ratios were estimated by quantification of the intensity of mutant undigested PCR products and comparing these to intensity of the digested product (Supp. Figure S2). For that purpose, ImageJ software was used (NIH, Bethesda, MD). Restriction digestion of each individual’s sample was performed in duplicate. PCR conditions and primers are available upon request.
Western Blot Analysis
Primary fibroblasts were lysed in 2% SDS lysis buffer, subjected to SDS-PAGE and transferred onto the PVDF membrane. SLC35A2 protein was detected using rabbit anti-SLC35A2 primary (Novus Biologicals, Littleton, CO) and goat anti-rabbit secondary antibodies (SeraCare Life Sciences Inc., Milford, MA). α-tubulin was detected with mouse primary (Developmental Studies Hybridoma Bank, Iowa City, IA) and sheep anti-mouse secondary antibodies (GE Healthcare, Chicago, IL). Western Blots were performed as previously described (Sosicka et al., 2017).
Immunofluorescence Staining
Endogenous UDP-galactose transporter was detected using anti-SLC35A2 primary (Novus Biologicals, Littleton, CO) and goat anti-rabbit Alexa Fluor 568 secondary (Life Technologies, Carlsbad, CA) antibodies diluted 1:400 and 1:200, respectively. Overexpressed SLC35A2 protein was counterstained with mouse anti-HA tag (Thermo Fisher Scientific, Waltham, MA) primary and goat anti-mouse Alexa-Fluor 568 secondary (Life Technologies, Carlsbad, CA) antibodies diluted 1:500 and 1:200, respectively. Terminal N-acetylgalactosamine (GalNAc) in O-glycans was detected either using Vicia villosa lectin (VVL) conjugated with FITC (Vector Biolabs, Malvern, PA) diluted 1:100 (fibroblasts) or biotinylated VVL lectin (Vector Biolabs, Malvern, PA) and streptavidin conjugated with Cy3 (Sigma Aldrich, Saint Louis, MO) both diluted 1:100 (CHO cells). Nuclei were counterstained using DAPI (MP Biomedicals, Santa Ana, CA). Immunofluorescence staining was performed as previously described (Sosicka et al., 2017). CHO-Lec8 cells were imaged using Zeiss LSM510 confocal microscope and fibroblasts with Zeiss LSM710 confocal microscope (Carl Zeiss, Oberkochen, Germany).
Generating CHO-Lec8 Stable Clones
Introduction of each SLC35A2 variant was carried out using QuikChange Site-Directed Mutagenesis Kit according to the manufacturer’s instruction (Agilent Technologies, Santa Clara, CA). PCR conditions as well as primer sequences are available upon request. Parental CHO and the Slc35a2 null CHO-Lec8 cells were grown in F-12K complete medium and transfected with respective pSelect-Zeo plasmid encoding either wild-type or mutant SLC35A2 with a HA epitope at the N-terminus, as previously described (Sosicka, Jakimowicz, Olczak & Olczak, 2014). Stable transfectants were generated by culturing cells as mentioned above supplemented with 200 μg/ml Zeocin for three weeks.
RESULTS
Variant Analysis
The majority of previously reported SLC35A2-CDG individuals, 27 of 32 (84%) (Supp. Table S1), were identified by NGS with many of those affected individuals listed in the online supplemental data of large sequencing studies (Ng et al., 2013; Kodera et al., 2013; EuroEPINOMICS-RES Consortium, Epilepsy Phenome/Genome Project & Epi4K Consortium, 2014; Dorre et al., 2015; Bosch et al., 2016; Lelieveld et al., 2016; Kimizu et al., 2017; Bruneel et al., 2018; Westenfield et al., 2018; Winawer et al., 2018; Yates et al., 2018; Sim et al., 2018). This explains why the number of reported subjects cited varies among studies. A useful online tool that helps to catalogue some publications involving de novo variants is http://denovo-db.gs.washington.edu/denovo-db/.
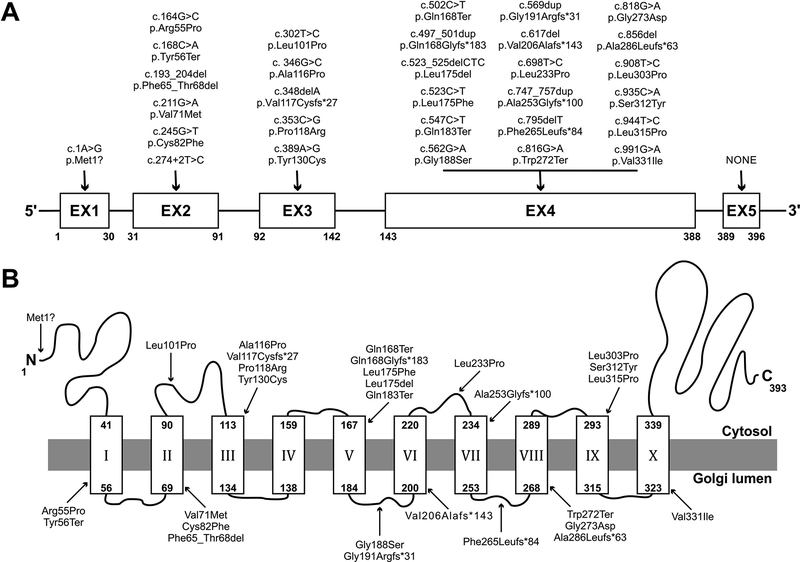
In our cohort, 29 of 30 of affected individuals (97%) were identified by NGS (Table 1). The one exception, CDG-0389, had an extremely abnormal CDT result suggestive of a galactosylation defect, which in combination with her clinical phenotype was suggestive of SLC35A2-CDG. Sanger sequencing confirmed the presence of a de novo c.698T>C [p.Leu233Pro] change (Table 1). In total, 26/30 (87%) were de novo variants not previously reported, while a few were recurrent de novo variants p.Arg55Pro, p.Gln168Ter, p.Ala253Glyfs*100 and p.Val331Ile (Ng et al., 2013; EuroEPINOMICS-RES Consortium, Epilepsy Phenome/Genome Project & Epi4K Consortium, 2014; Westenfield et al., 2018; Winawer et al., 2018; Sim et al., 2018) (Table 1, Fig. 1). CDG-0468 carried the c.502C>T [p.Gln168Ter] and was part of the Epi4K study mentioned above. The distribution of the 30 variants included 15 missense, 7 out-of-frame INDELS, 4 nonsense, 2 in-frame deletions, 1 loss of an essential splice site and 1 start codon loss (Table 1, Fig. 1). Only one variant, p.Gly188Ser, was present at very low frequency (2/133432 alleles) in the gnomAD database (http://gnomad.broadinstitute.org/) (gnomAD r2.0.2 accessed 1.24.2019) of 125,748 exomes and 15,708 whole-genomes of unrelated individuals. We previously reported that SLC35A2 does not tolerate genetic variation, especially in males (Ng et al., 2013). Consistent with that observation, the gender ratio was 29 females to 1 male (Table 1). The single male (CDG-0460) identified in this study did not appear to be mosaic for the c.944T>C (p.Leu315Pro) based on his NGS allele reads, which only showed the presence of the mutant allele in tested blood samples, no other tissues were studied (data not shown). It should be noted that complete variant segregation was unavailable for two families. CDG-0114 carried a novel c.617del [p.Val206Alafs*143] and since it is generally accepted that SLC35A2 does not tolerate INDEL variants, it is unlikely the father, who was unavailable for testing, was a carrier. CDG-0248 carried a novel missense variant c.818G>A [p.Gly273Asp] that was absent from the mother, but the father was unavailable for testing. However, in a recent separate study on SLC35A2-CDG, an unrelated affected individual was identified carrying the same c.818G>A [p.Gly273Asp] as a de novo variant showing a slightly abnormal CDT (Vals et al., 2019 doi: 10.1002/jimd.12055)
Table 1. General information for 30 individuals with SLC35A2-CDG.
Individual ID, gender, genotypes, predicted CADD scores, CDT results and detection method are provided for the 30 individuals found to carry de novo variants within SLC35A2. Nucleotide numbering for cDNA uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1. SLC35A2 NCBI Accession (NM_001042498.2) and for ENSEMBL (ENST00000376521.6). All cDNA to protein translations were confirmed using https://mutalyzer.nl/ (WES – Whole Exome Sequencing, WGS – Whole Genome Sequencing) N/A – Not Available
| ID | Sex | cDNA Position | Protein Position | Variant Status | CADD Score | CDT Result | Detection Method |
|---|---|---|---|---|---|---|---|
| CDG - 0008 | F | c.523_525del | p.Leu175del | Novel | N/A | N/A | WES |
| CDG - 0028 | F | c.193_204del | p.Phe65_Thr68del | Novel | N/A | Normal | WES |
| CDG - 0045 | F | c.816G>A | p.Trp272Ter | Novel | 38 | Normal | Panel |
| CDG - 0052 | F | c.348del | p.Val117Cysfs*27 | Novel | N/A | Normal | WES |
| CDG - 0056 | F | c.562G>A | p.Gly188Ser | Novel | 8.6 | N/A | WES |
| CDG - 0057 | F | c.497_501dup | p.Gln168Glyfs*183 | Novel | N/A | N/A | Panel |
| CDG - 0058 | F | c.168C>A | p.Tyr56Ter | Novel | 22.2 | Normal | Panel |
| CDG - 0059 | F | c.274+2T>C | p.? | Novel | N/A | Normal | WES |
| CDG - 0114 | F | c.617del | p.Val206Alafs*143 | Novel | N/A | Normal | WGS |
| CDG - 0115 | F | c.856del | p.Ala286Leufs*63 | Novel | N/A | N/A | WGS |
| CDG - 0116 | F | c.547C>T | p.Gln183Ter | Novel | 36 | Normal | Panel |
| CDG - 0149 | F | c.908T>C | p.Leu303Pro | Novel | 31 | Normal | WES |
| CDG - 0154 | F | c.795del | p.Phe265Leufs*84 | Novel | N/A | Normal | WES |
| CDG - 0164 | F | c.935C>A | p.Ser312Tyr | Novel | 26.6 | Normal | WES |
| CDG - 0173 | F | c.991G>A | p.Val331IIe | Known | 26.4 | Normal | WES |
| CDG - 0187 | F | c.211G>A | p.Val71Met | Novel | 26.9 | Normal | WGS |
| CDG - 0198 | F | c.346G>C | p.Ala116Pro | Novel | 26.6 | N/A | Panel |
| CDG - 0199 | F | c.302T>C | p.Leu101Pro | Novel | 26.1 | Normal | WES |
| CDG - 0248 | F | c.818G>A | p.Gly273Asp | Novel | 23.3 | Normal | WES |
| CDG - 0265 | F | c.353C>G | p.Pro118Arg | Novel | 27 | Normal | Panel |
| CDG - 0266 | F | c.747_757dup | p.Ala253Glyfs*100 | Known | N/A | Normal | Panel |
| CDG - 0308 | F | c.523C>T | p.Leu175Phe | Novel | 26.6 | Abnormal | Panel |
| CDG - 0332 | F | c.164G>C | p.Arg55Pro | Known | 26.7 | Normal | WES |
| CDG - 0389 | F | c.698T>C | p.Leu233Pro | Novel | 27.8 | Abnormal | Sanger |
| CDG - 0449 | F | c.1A>G | p.Met1? | Novel | 23.2 | Normal | WGS |
| CDG - 0454 | F | c.569dup | p.Gly191Argfs*31 | Novel | N/A | Normal | WES |
| CDG - 0460 | M | c.944T>C | p.Leu315Pro | Novel | 27.4 | Abnormal | WES |
| CDG - 0468 | F | c.502C>T | p.Gln168Ter | Known | 36 | Normal | WES |
| CDG - 0469 | F | c.389A>G | p.Tyr130Cys | Novel | 28.1 | Abnormal | WES |
| CDG - 1039 | F | c.245G>T | p.Cys82Phe | Novel | 28.5 | Normal | WES |
Figure 1.
Schematic representation of the SLC35A2 variants present in our 30 individuals. (A) Schematic representation showing exon location of all 30 SLC35A2 variants identified in this study. (B) Schematic representation showing protein localization of all 30 SLC35A2 variants identified in this study. SLC35A2 topology was determined using 3D model generated with PHYRE2 server (Kelley, Mezulis, Yates, Wass & Sternberg, 2015).
The gnomAD database for the canonical transcript of SLC35A2 (ENST00000247138) contains no hemizygous or heterozygous variants causing truncation of the protein including no loss of a start codon, premature stop codons or out-of-frame INDELS, or in frame deletions upstream of the C-terminal cytoplasmic tail. The database does contain several in frame deletions within the cytoplasmic tail that do not disrupt transmembrane domain (TMD) organization and so are unlikely to disrupt SLC35A2 activity: p.Ala355_Ser356dup (n=17), p.Ala355_Ser356del (n=1), p.Ser356_Gly357del (Sosicka, Jakimowicz, Olczak & Olczak, 2014).
In silico modeling to predict the deleterious nature of a specific variant was performed using the Combined Annotation Dependent Depletion (CADD) (http://cadd.gs.washington.edu/) scoring method version GRCh37-v1.4 (Kircher et al., 2014). This method was solely used as a guide to aid in prioritizing variants that would be tested in our complementation assay. Using the CADD method, all (nonsense or missense) but one variant scored ≥22, placing it within the top 1% of predicted deleterious variants (Table 1). Further, sixteen (80%) had CADD scores (>25) in the top 0.5% of predicted deleterious variants (Table 1).
Clinical Summary
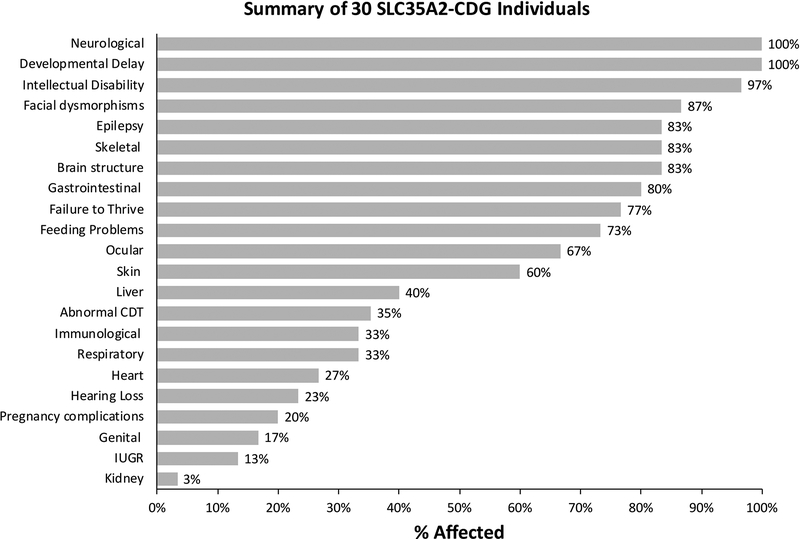
In total, we obtained clinical data on 30 affected individuals (29 females/1 male) from 30 unrelated families. Twenty-eight individuals were confirmed by either Sanger sequencing or NGS analysis of the trio, to harbor de novo variants in SLC35A2 while the remaining two are strongly suggestive to be de novo as mentioned above. In our cohort, 6/30 (20%) (Fig. 2) pregnancies experienced some type of fetal anomaly with two possible skeletal dysplasias and two with pericardial effusions at 20 weeks that spontaneously resolved by 30 weeks. Individual CDG-0056 had decreased fetal movements and a breech presentation. Feeding problems were seen in 22/30 (73%) individuals with 20/30 (67%) having placement of a g-tube. Consequently, 23/30 (77%) affected individuals displayed clear features of failure to thrive (Fig. 2).
Figure 2.
Clinical summary of 30 individuals with de novo variants in SLC35A2. Clinical information for each of the 30 individuals were provided by clinicians and summarized as a percentage of affected individuals.
For many CDG types that affect N-linked glycosylation, serum transferrin is a useful and reliable tool for the initial screening of CDG. Abnormal N-glycans associated with SLC35A2-CDG typically display a truncated pattern lacking galactose and terminal sialic acid (Ng et al., 2013; Dorre et al., 2015). However, this is not always the case for SLC35A2-CDG. In fact, of the thirty-two previously reported subjects, only five (16%) had abnormal CDT results consistent with SLC35A2-CDG (Ng et al., 2013; Dorre et al., 2015; Bruneel et al., 2018) (Supp. Table S1). Adding further complexity, at least two of these individuals showed complete normalization of their CDT by three years of age without any specific therapy (Ng et al., 2013). Since many SLC35A2-CDG are not diagnosed until after the age of three, it begs the question how many of the previously reported subjects may have normalized their CDT.
In our cohort, 5/21 (24%) individuals who had CDT analysis, showed abnormal results consistent with SLC35A2-CDG (Fig. 2). Importantly, among those, three are >3 years of age and one (CDG-0469) has shown spontaneous improvement in her CDT without clinical improvement (Supp. Figure S3). While the CDT profile for CDG-0469 has markedly improved, the N-glycans on her serum IgG have remained normal over the last several years (data not shown). The remaining two are still under the age of three, but repeated CDTs have been consistently abnormal thus far. Among 21 individuals tested for CDT, 17 employed mass spectrometry.
Similar to previously reported subjects, our cohort of SLC35A2-CDG individuals had varying degrees of physical and neurodevelopmental deficiencies including developmental delay (30/30–100%), epilepsy (25/30–83%), hypotonia (28/30–93%), microcephaly (13/30–43%) and abnormal brain imaging results (25/30–83%) (Fig. 2). White matter abnormalities 16/30 (53%) and cerebellar atrophy 17/30 (57%) comprised the majority of the abnormal brain imaging results (Supp. Table S2). Intellectual disability was present in 28/29 (97%) individuals who could be evaluated (Fig. 2). Another major clinical feature was a recognizable skeletal abnormality, present in 25/30 (83%). Individuals with skeletal problems usually presented with multiple features including shortened limbs (9/30–30%), contractures (8/30–27%), scoliosis (15/30–50%) and hand or finger abnormalities (16/30–53%) (Supp. Table S2). These individuals may have shown a greater impairment of proteoglycan synthesis, which is critical for normal bone development or perhaps a greater degree of SLC35A2 mosaicism. Facial dysmorphism was seen in 26/30 (87%) individuals, while ocular abnormalities comprised 20/30 (67%) (Fig. 2). Cortical visual impairment (CVI) was the most frequent ocular abnormality, reported in 13/30 (43%) (Supp. Table S2). The main skin finding was inverted nipples 8/30 (27%), although 4/30 (13%) individuals had unusually pigmented areas of skin (Supp. Table S2).
Less affected systems included the heart 8/30 (27%) and kidney 1/30 (3%), while liver involvement (mildly elevated transaminases) was noted in 12/30 (40%) individuals (Fig. 2) (Supp. Table S2). Four affected females showed premature signs of puberty. Interestingly, sensorineural hearing loss, while not often seen in other forms of CDG was found in three subjects (Supp. Table S2).
Finally, SLC35A2-CDG appears to have a low mortality rate. No fatalities were seen in the 32 previously reported individuals and in our group of 30, all but one is still alive. In the most prevalent type of CDG, PMM2-CDG, 20% of affected individuals die within the first year of life (de Lonlay et al., 2001; Grunewald, 2009).
Optimization of UDP-Galactose Transport Assay
SLC35A2 is known to be a core component of glycosylation, especially for N- and O-linked, glycosaminoglycan (GAG) and glycolipid synthesis which are heavily reliant on galactosylation (Miura et al., 1996; Toma, Pinhal, Dietrich, Nader & Hirschberg, 1996; Brandli, Hansson, Rodriguez-Boulan, Simons, 1998). Yet, an unusual biochemical hallmark of SLC35A2-CDG is that most affected individuals have either normal CDT or normal total N- and O-linked glycan compositions (Xia et al., 2013) Since CDT may not be an effective screening tool for this particular CDG, many cases with SLC35A2-CDG are less likely to be diagnosed. Of the 32 reported SLC35A2-CDG individuals, only the original three subjects had direct measurement of UDP-galactose transport assayed using primary fibroblasts (Ng et al., 2013). Therefore, we set out to develop a robust and reliable biochemical assay for measuring SLC35A2-dependent Golgi transport.
Glycosides are small, membrane-permeable molecules consisting of a hydrophobic aglycon and a monosaccharide. When localized in the proper Golgi compartment, glycosides in close proximity to their respective glycosyltransferase can serve as primers for oligosaccharide biosynthesis (Sarkar, Fritz, Taylor & Esko, 1995; Sarkar, Rostand, Jain, Matta & Esko, 1997). For this reason, glycosides were used to study co-localization of glycosyltransferases and nucleotide sugar transporters (Etchison, Srikrishna & Freeze, 1995; Etchison & Freeze, 1996; Portner, Etchison, Sampath & Freeze, 1996; Kim, Miura, Etchison & Freeze, 2001). We used glycoside GlcNAcβ−4-MU as an artificial acceptor to measure SLC35A2-dependent UDP-galactose transport into the Golgi. Specifically, we hypotonically-lysed cells and added membrane permeant GlcNAcβ−4-MU to load intact Golgi with the glycoside acceptor. When exogenous UDP-[6-3H] galactose is provided, it is transported by SLC35A2 into the Golgi where it becomes accessible to the co-located β1,4 galactosyltransferase (β1,4GALT). The colocalization of donor (transported UDP-Gal), acceptor glycoside and transferase within the Golgi allows the transported UDP-[6-3H] galactose to be transferred to GlcNAcβ−4-MU. The final reaction product, [6-3H] Galβ1,4GlcNAcβ−4-MU, may also diffuse back across the membranes, but much more slowly (Etchison, Srikrishna & Freeze, 1995). Furthermore, [6-3H] Galβ1,4GlcNAcβ−4-MU can be extended with a terminal sialic acid (Etchison, Srikrishna & Freeze, 1995). The great majority of galactosylated glycoside remains inside the Golgi during the assay and is extracted with 70% ethanol.
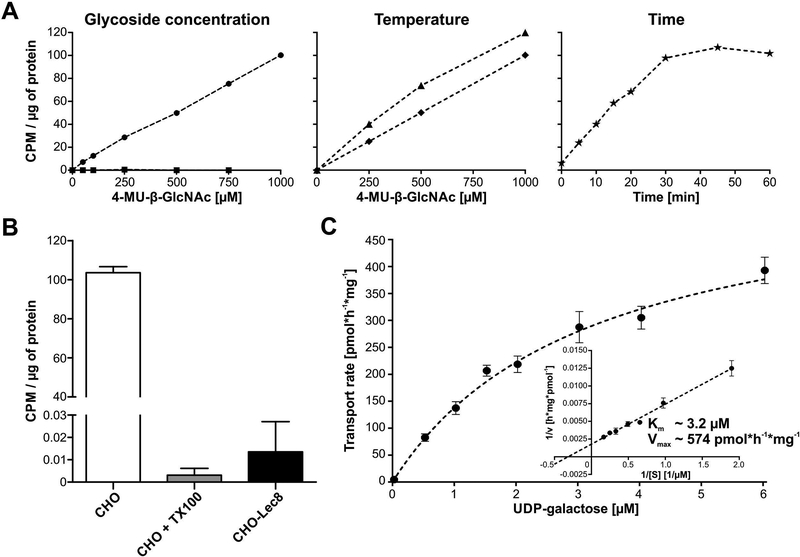
We optimized assay conditions using CHO and CHO-Lec8 cells. CHO-Lec8 is a well- characterized SLC35A2 mutant, which does not produce a functional UDP-galactose transporter and thus is incapable of delivering this the donor into the Golgi (Oelmann, Stanley & Gerardy-Schahn, 2001). In this study we used the CHO-Lec8 line (Stanley, 1981) carrying a 101bp deletion which results in a premature stop codon.
We optimized GlcNAcβ−4-MU concentration, reaction temperature and time (Fig. 3A) for permeabilized cells, which were pre-incubated with glycoside for 5 minutes prior to adding UDP-[6-3H] galactose. Glycoside concentrations were selected (Etchison, Srikrishna & Freeze, 1995; Etchison & Freeze, 1996; Portner, Etchison, Sampath & Freeze, 1996; Kim, Miura, Etchison & Freeze, 2001) based on formation of [6-3H] Galβ1,4GlcNAcβ−4-MU. CHO-Lec8, showed no activity confirming specificity of the assay (Fig. 3A) and 37°C was the optimal temperature (Fig. 3A) with an incubation time of 45–60 minutes (Fig. 3A). To ensure the UDP-[6-3H] galactose incorporation is SLC35A2-dependent, we performed the assay on CHO cells in the presence of 0.025% Triton X-100, which is slightly above the critical micelle concentration (CMC) and will disrupt the Golgi membrane integrity and therefore dramatically decrease labeling of GlcNAcβ−4-MU in the Golgi lumen (Hayes, Freeze & Varki, 1993). The detergent abolished all transport (Fig. 3B), proving the approach requires membrane integrity and the colocalization of donor, acceptor substrates and glycosyltransferase. Glycoside products were verified by HPLC as described (Etchison, Srikrishna & Freeze, 1995), showing 80% [6-3H] Galβ1,4GlcNAcβ−4-MU and 20% extended by addition of 2,3 linked sialic acid (data not shown).
Figure 3.
Optimization of the glycoside-based UDP-galactose transport assay. (A) GlcNAcβ−4-MU concentration, temperature and time of the assay were optimized. Glycoside concentration was tested using CHO (circles) and CHO-Lec8 (squares) cells. Temperature and time were optimized only on CHO lines. Transport efficiency at room temperature (diamonds) and 37 °C (triangles) was compared using increasing concentrations of GlcNAcβ−4-MU. Time curve (asterisk) was assayed at 37°C with 1 mM glycoside. (B) UDP-galactose transport was measured in CHO cells using optimized conditions. As a negative control CHO-Lec8 cells were employed. Specificity of the assay was determined by treating CHO cells with 0.025% Trition-X100 (CHO + TX100). The assay was performed in three biological repetitions. Error bars represent SD (C) Michaelis constant and the maximum UDP-galactose transport velocity were determined for CHO protein in optimized assay conditions. Each point represents a mean of 3 to 6 independent biological repetitions using different cell preparations and performed on different days. Error bars represents SEM. CPM – Counts Per Minute
Finally, we showed that this approach can be used to determine the kinetics of UDP-galactose transport in CHO cells. Under optimal conditions we determined the transporter Km as ~3.2 μM Vmax of ~574 pmol UDP-galactose/h/mg total protein in the cell lysate (Fig. 3C).
UDP-Galactose Transport in Fibroblasts
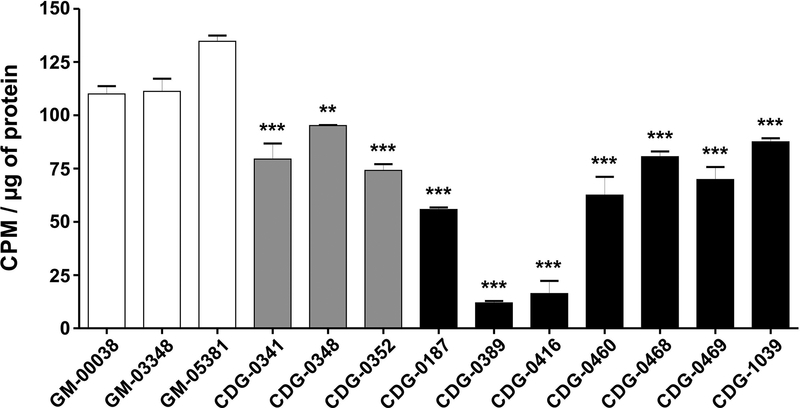
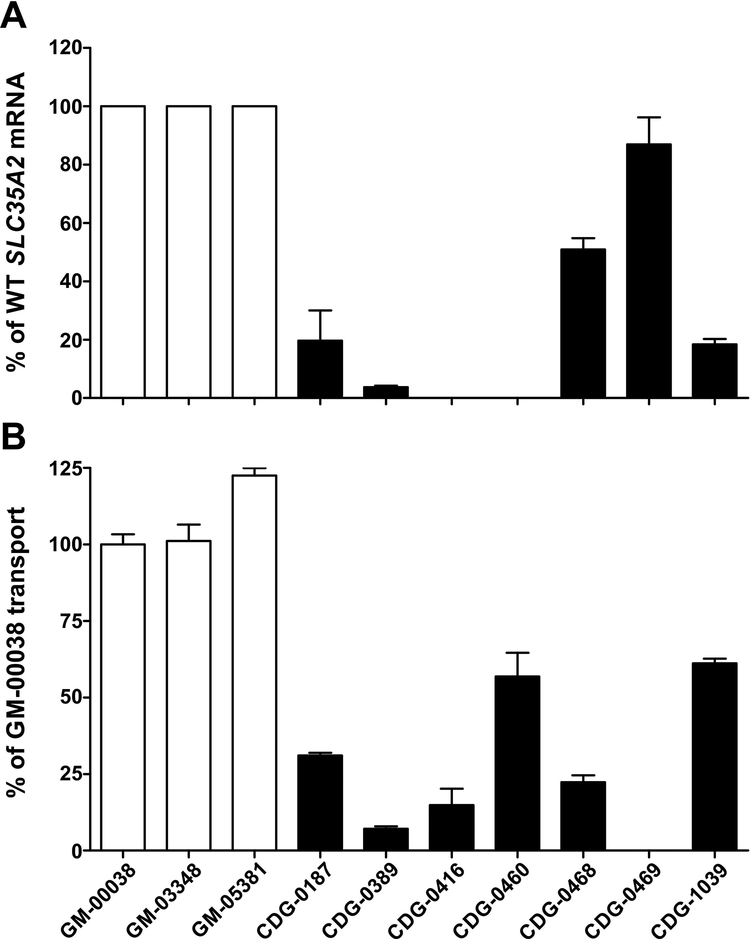
We initially measured UDP-galactose transport in three commercially available control fibroblast lines from apparently healthy individuals (Coriell cell repository GM-00038, GM-03348, GM-05381). Subsequently, we assayed ten primary fibroblast lines from individuals with confirmed de novo variants in SLC35A2, including three previously characterized SLC35A2-CDG subjects. Importantly, each SLC35A2-CDG-derived cell line showed varying levels of statistically significant reductions of UDP-[6-3H] galactose transport, while the three controls showed consistently comparable levels (Fig. 4). Two individuals had nearly no transport activity (CDG-0389, 0416) (Fig. 4). CDG-0416 is an affected male from another study for whom we only have biochemical results. However, it is important to note that this individual, like CDG-0460, only expressed the mutant allele. Comparing results from the first three reported individuals, using the transport method by Ng et.al to those presented here, yielded comparable results (Fig. 4) (Ng et al., 2013). We further characterized these fibroblast lines to better explain these differences in SLC35A2-dependent transport activity.
Figure 4.
UDP-galactose transport assay in primary fibroblasts. UDP-[6-3H] galactose transport in permeabilized fibroblast cells with intact Golgi apparatus under optimized conditions. Each assay was performed in three biological repetitions and expressed as a percent of the control (* p < 0.05; ** p < 0.005; *** p<0.001).
SLC35A2 cDNA Allele Ratios
We had previously shown in mosaic males that wild-type fibroblasts grew faster than those containing SLC35A2 pathogenic variants. Since females could have variable proportions of X-inactivated cells, it was important to assess the proportions of normal and variant alleles. To estimate the ratio of wild-type and mutant alleles, we took advantage of the fact that each variant creates either a gain or loss of a particular restriction site (Supp. Figure S1; Supp. Table S3). RNA isolated from control and SLC35A2-CDG fibroblasts was transcribed into cDNA, and PCR amplified and digested with the appropriate restriction enzyme (Supp. Table S3). We found that CDG-0416 and CDG-0460 expressed only the mutant SLC35A2 allele (Fig. 5A), which was consistent with the NGS data. In the other five individuals, we found ~ 4% (CDG-0389), 20% (CDG-0187), 51% (CDG-0468), 87% (CDG-0469) and 18% (CDG-1039) of the wild-type mRNA remaining (Fig. 5A). This observation, can in part, explain the differences in UDP-galactose transport activity from each subject. Moreover, knowing the ratio of wild-type to mutant alleles allowed us to approximate the contribution of the specific variants on SLC35A2-dependent transport activity. However, it should be noted that allele ratios are likely to vary in different tissues for those individuals and could also change over time. For example, the variable allele ratio in the liver can provide a possible explanation for why many SLC35A2-CDG individuals have normal CDT results. Over time, if mutant liver cells are selected against, the majority will be normal cells.
Figure 5.
Determination of the wild-type to mutant allele ratios in fibroblasts (A) SLC35A2 cDNA allele ratios in primary fibroblasts from control and SLC35A2-CDG subjects. Wild-type to mutant allele ratios were determined using a restriction enzyme-based assay. GM-00038, GM-03348 and GM-05381 cells were used as controls. Each assay was performed in duplicate in order to determine reproducibility of the assay. (B) UDP-galactose transporter activity was calculated based on allele ratio presented as a percentage of GM-00038 transport activity. To illustrate the activity of the respective mutant protein, the contribution of wild-type protein was subtracted.
Based on the measured ratios of wild-type to mutant alleles, we attempted to estimate the residual transport activity for each SLC35A2 protein variant. GM-00038 transport activity was taken as a reference for the three controls. We found that the UDP-galactose transporter mutant present in CDG-0187 cells exhibits ~30%, CDG-0389 ~7%, CDG-0416 ~15%, CDG-0460 ~55%, CDG-0468 ~20%, CDG-0469 0% and in CDG-1039 ~60% of wild-type protein activity (Fig. 5B). However, care must be taken when interpreting these values. For example, one control line (GM-05381) exhibited significantly higher transport activity than the others tested reaching ~125% of GM-00038 transport efficiency. This indicates that the transport yield may differ between control lines. For these reasons, these values are a relative comparison of activity in the presence of a particular allele ratio.
Furthermore, X-inactivation can differ between tissues and allele ratio may change with increased passage of fibroblast cell lines. In our cohort, only 4 of 29 female individuals had XCI studies performed with two being reported as having random XCI, while the other two showed non-random (i.e “skewed”) X-chromosome inactivation (data not shown). Importantly, CDG-0389 had XCI analysis done from three tissue sources; buccal cells, fibroblasts and whole blood using a previously reported method (Allen et al., 1992). Non-random inactivation was seen in both buccal (94:6) and cultured fibroblasts (85:15), while blood samples (57:42) appeared to be random. Ratios greater than 80:20 are considered to be indicative of Non-random X inactivation.
Western Blot Analysis
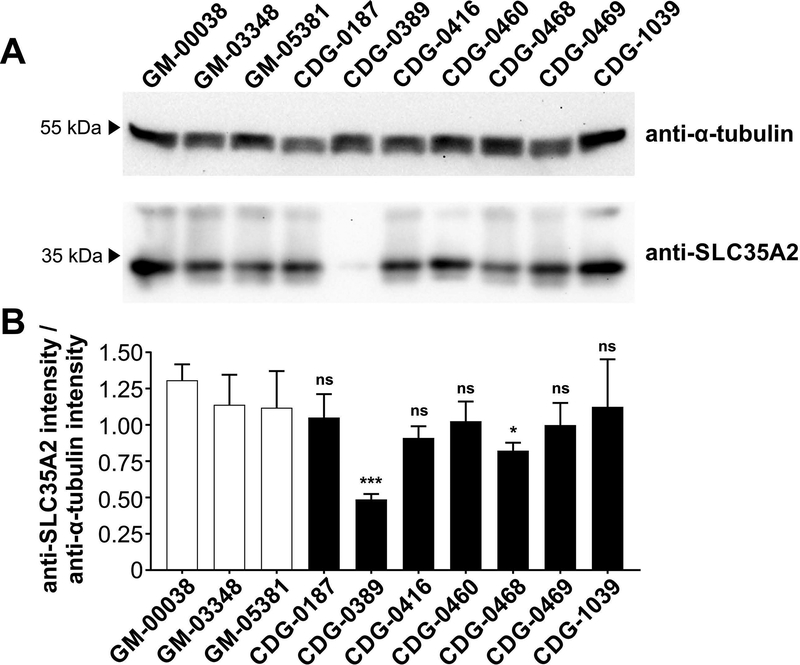
Primary fibroblasts were used for western blot analysis of SLC35A2 protein from seven affected individuals to determine whether the decrease in UDP-galactose transport could be attributed to a decrease in SLC35A2 protein. We found that CDG-0389, who carriers c.698T>C (p.Leu233Pro), had no detectable SLC35A2 protein, while a slight decrease was observed in CDG-0468, who harbors a premature stop codon c.502C>T (p.Gln168Ter) (Fig. 6A, Table 1). In the other five subjects, SLC35A2 protein levels were not affected by the de novo variant (Fig. 6A). Statistical significance was calculated on multiple blots run on separate biological replicates (Fig. 6B)
Figure 6.
Western Blot analysis of SLC35A2 protein from primary fibroblast. (A) Western Blot analysis of SLC35A2 protein from a total of seven SLC35A2-CDG fibroblast lines as well as three controls (GM-00038, GM-03348 and GM-05381). Anti-α-tubulin antibody was used as a loading control. (B) Quantification was performed by normalizing SLC35A2 to α-tubulin from four independent blots using two biological replicates collected on separate days with error bars calculated as a StdDev.
Immunofluorescence Staining of Fibroblasts
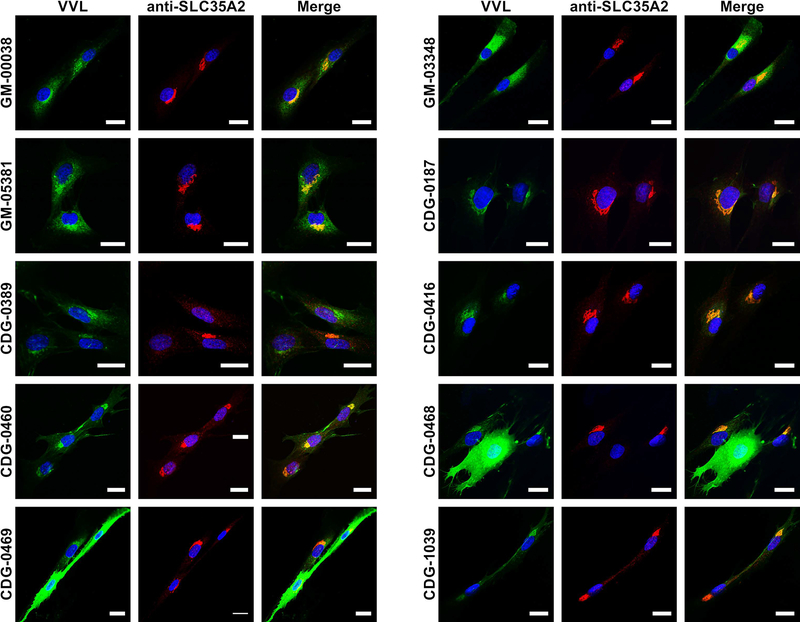
Fibroblasts from both controls and subjects were seeded onto coverslips, grown for 48 hours and stained using anti-SLC35A2 antibody and VVL lectin. This lectin specifically recognizes terminal GalNAc present in O-glycans, which is usually masked by addition of galactose eliminating lectin binding. Since VVL is an established marker, which exhibits significantly higher amount in cells with UDP-galactose transporter defects (Maszczak-Seneczko, Olczak, Wunderlich & Olczak, 2011, Ng et al., 2013), we expected to see elevated VVL reactivity in fibroblasts from the subjects. Confocal microscopy confirmed that SLC35A2 protein properly localized to the Golgi apparatus in all CDG cells (Fig. 7), but the predicted increase in VVL binding was observed in only two of seven SLC35A2-CDG fibroblasts (CDG-0468 and CDG-0469). The other cells exhibit similar staining pattern and fluorescence intensity as the control lines. There are several possible explanations for this effect. First, the overall biosynthesis of O-glycans might be significantly depleted in these cells leading to reduction in terminal GalNAc. Second, other monosaccharides can be attached to GalNAc, blocking VVL binding. Finally, O-glycans might not be a representative marker of a galactosylation defect in all individuals, while N-glycans, proteoglycans or glycolipids could be better in some than others.
Figure 7.
Immunofluorescence staining of primary fibroblasts. To assess glycosylation between control (GM-00038, GM-03348 and GM-05381) and subject fibroblasts (CDG-0187, CDG-0389, CDG-0416, CDG-0460, CDG-0468, CDG-0469 and CDG-1039) VVL lectin was used (green). This lectin specifically recognizes terminal GalNAc present in O-glycans, which is usually masked by galactose and blocks the lectin reactivity. UDP-galactose transporter was counterstained with anti-SLC35A2 antibody (red). Scale bar 20 μm.
Western blot results revealed that both CDG-0389 and CDG-0468 produced significantly reduced levels of the SLC35A2 protein (Fig. 6A), which was consistent with the immunofluorescence results (Fig. 7).
CHO Lec8 Complementation Assays
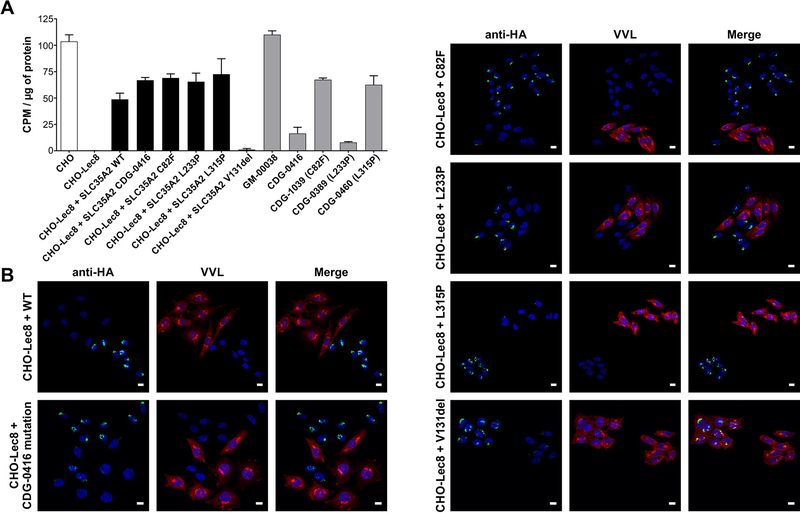
We set out to determine if the CHO mutant cell line Lec8, which lacks all galactosylation, could be used for a cell-based complementation assay. We expected that only wild-type or benign SLC35A2 variants would restore UDP-galactose transport and galactosylation in the mutant while expressing a pathogenic variant would dramatically lower transport and galactosylation. We generated five stable Lec8 lines expressing wild type or specific SLC35A2 variants for which we also had primary fibroblasts for comparison. Expression of these variants were driven off either weak or strong promoters. The first method was the UDP-galactose transport assay and the second was the immunofluorescence staining with VVL lectin. In the transport assay, wild-type SLC35A2 was able to correct the transport defect in Lec8. However, the p.Cys82Phe, p.Leu233Pro, p.Leu315Pro and the protein change present in CDG-0416 were also able to complement the Lec8 defect (Fig. 8A). In contrast, these results were not seen in fibroblasts from individuals with the same SLC35A2 variants (Fig. 4).
Figure 8.
Characterization of CHO-Lec8 stable clones expressing SLC35A2. (A) UDP-[6-3H] galactose transport assay in parental CHO, CHO-Lec8 and CHO-Lec8 cells overexpressing respective variants of SLC35A2 protein. For better comparison, fibroblast results from control (GM-00038) and four affected individuals (CDG-0389, CDG-0416, CDG-0460, CDG-1039) were plotted. The transport for SLC35A2-CDG fibroblasts was expressed as the predicted activity of mutant transporter by subtracting the contribution of wild type protein, based on the calculated allele ratio. Each assay was performed in two biological repetitions. (B) Immunofluorescence staining of CHO-Lec8 cells overexpressing UDP-galactose transporter. Overexpressed HA-tagged SLC35A2 protein was detected with anti-HA antibody (green). VVL lectin staining was used to detect glycosylation changes between CHO-Lec8 cells which do or do not overexpress the transporter (red). Stable transfectants, but not clonal populations were analyzed. Scale bar 20 μm.
Next, we tested if stably transfected cells had reduced VVL lectin reactivity due to the masking of the VVL epitope by galactose. While the tested variants localized properly to the Golgi apparatus (Fig. 8B), all were equal to wild-type in correcting the galactosylation defect (Fig. 8B). This cannot be true since these mutants are not capable of transporting UDP-galactose as seen using the primary fibroblast. Only overexpression of a loss of function variant, p.Val131del, did not correct CHO-Lec8 phenotype (Fig. 8). The data suggest that simply over-expressing SLC35A2 variants in Lec8 or any cell line could have unforeseen consequences and not be truly reflective of physiological conditions seen in fibroblast from affected individuals cells.
Biochemical Summary
We characterized seven affected individuals and demonstrate that currently only subject-derived fibroblasts provide a valid method to measure UDP-galactose transport to confirm pathogenicity of the variants (Fig. 4; Table 2). Only 2 of 7 lines (CDG-0389 and CDG-0468) produced significantly decreased amount of SLC35A2 protein (Figs 6 and 7; Table 2). In contrast, the other hallmark expected to be present in all SLC35A2-CDG cells is increased binding of VVL to terminal GalNAc, however, this feature was found only in 2 out of 7 fibroblasts (CDG-0468 and CDG-0469; Fig. 7; Table 2). It is noteworthy that the percentage of VVL positive cells assigned by immunofluorescence staining corresponds with the determined cDNA mutant to wild-type allele ratios (Supp. Figure S4; Table 2). Because of the limited access to fibroblasts from affected individuals, we tried to establish an alternative method, using complemented CHO-Lec8 cells. Attempts to make stable CHO-Lec8 cells expressing SLC35A2 driven off a CMV promoter often resulted in cell death. Using a weaker EF-1 promoter resulted in successful stable SLC35A2 expression, however we clearly show that overexpression of subject-specific variants resulted in a phenotype similar to that of the wild-type protein (Fig. 8; Table 2). Thus, CHO-Lec8 complemented cells are not a suitable diagnostic tool.
Table 2. Summary of biochemical characteristics from seven SLC35A2-CDG fibroblast lines.
Summary of characteristics from seven fibroblast lines including UDP-galactose transport, wild-type to mutant allele ratio, SLC35A2 protein level and VVL reactivity.
| Variant type | Decreased UDP-Gal transport in fibroblasts |
% expression of WT allele | % of WT transport activity | Decreased amount of SLC35A2 protein | Increased VVL reactivity | % of VVL negative cells | |
|---|---|---|---|---|---|---|---|
| CDG-0187 | missense | Yes | ~20% | ~31% | No | No | NA |
| CDG-0389 | missense | Yes | ~4% | ~7% | Yes | No | NA |
| CDG-0416 | missense | Yes | 0% | ~15% | No | No | NA |
| CDG-0460 | missense | Yes | 0% | ~57% | No | No | NA |
| CDG-0468 | nonsense | Yes | ~51% | ~20% | Yes | Yes | ~63% |
| CDG-0469 | missense | Yes | ~87% | 0% | No | Yes | ~83% |
| CDG-1039 | missense | Yes | ~18% | ~61% | No | No | NA |
DISCUSSION
Here we describe the identification and characterization of 30 individuals with SLC35A2-CDG, which to date is the single largest study for this rare disorder. As in previously reported SLC35A2-CDG subjects, all thirty individuals in our cohort had profound neurological and developmental impairments. Many (83%), but not all had epilepsy. We find a large percentage (83%) of individuals also displayed some form of skeletal abnormality, which has not been previously highlighted.
Since many affected individuals with SLC35A2-CDG do not show an abnormal CDT or abnormal total serum N- or O-glycan profile, we set out to establish a reliable and robust method for assaying SLC35A2-dependent transport activity in primary fibroblasts to characterize potential VUS. While sometimes difficult to obtain, primary fibroblasts allow the direct measurement of SLC35A2-dependent transport activity and wild-type to mutant allele ratios. The latter point could help explain why some individuals still retain significant transport activity in fibroblasts.
It would be ideal to develop additional non-fibroblast lines for verifying subject-relevant variants or reclassifying VUS. Early work establishing the role for SLC35A2 in galactosylation had been primarily performed using non-human mutant cell lines from murine mammary carcinoma cells (FM3A), Chinese hamster ovary (CHO) cells and Madin-Darby Canine Kidney (MDCK) cells (Stanley, 1981; Ishida et al., 1999; Oelmann, Stanley & Gerardy-Schahn, 2001; Olczak & Guillen, 2006). We attempted to stably express subject-specific SLC35A2 variants in CHO-Lec8 cells but could not achieve consistent results in comparison to fibroblasts from SLC35A2-CDG individuals, despite using expression vectors containing either weak or strong promoters.
Currently, only subject-derived fibroblasts provide a valid method to measure UDP-galactose transport to confirm pathogenicity of the variants. We show by immunofluorescence staining that resident glycans may not always lead to a clear conclusion and may partially explain why a large group of affected individuals do not exhibit any changes in transferrin glycosylation. We have established here that CHO-Lec8 cells, which do not produce their own functional UDP-galactose transporter, cannot be used for the purpose of complementation. All tested fibroblasts exhibited impaired UDP-galactose transport; however, when selected variants were overexpressed in CHO-Lec8 cells, the respective proteins appeared fully functional (Fig. 8). We believe that using complemented cell lines to assay SLC35A2 activity is not a suitable approach. In the end, if fibroblast analysis is not possible, generating individualized CRISPR/Cas9 edited cell lines might be needed for accurate characterization of specific variants.
To date only 5/32 previously reported individuals had abnormal transferrin glycosylation (Ng et al., 2013; Dorre et al., 2015; Bruneel et al., 2018). Similarly, in our cohort 5/21 subjects had abnormal results that were consistent with SLC35A2-CDG. Analysis of SLC35A2-CDG fibroblasts also revealed that only two out of seven had impaired galactosylation (Fig. 7). It remains unclear why these lines do not show galactosylation deficiencies, when UDP-galactose transport measured in fibroblasts is clearly deficient. However, such a result is not unprecedented. CRISPR mediated SLC35A5 knock-out in HepG2 cells causes a significant decrease in UDP-GlcNAc, UDP-GalNAc and UDP-GlcA Golgi transport without affecting global glycosylation (Sosicka et al. 2019). Residual transport activity seen in fibroblasts (Fig. 4 and Fig. 5B) may be sufficient for normal glycosylation. Alternatively, another sugar nucleotide transporter might contribute to or compensate for decreased SLC35A2-dependent UDP-galactose transport. A possible candidate is the UDP-GlcNAc transporter, which can partially restore galactosylation when overexpressed in CHO-Lec8 and MDCK-RCAr cells (Maszczak-Seneczko, Olczak, Jakimowicz & Olczak, 2011b).
With a large number of mutations available, we wanted to describe the variants based on protein structural features. The previously reported missense variants, Arg55, Phe65, Ala170, Leu172, Val206, Ser213, Val331, and, of those reported here, Arg55, Val71, Cys82, Leu101, Ala116, Leu175, Gly188, Gly273, Leu315, Val331 appear to be localized within one of the ten hydrophobic transmembrane domains (TMD) predicted by UNIPROT, suggesting that mutations disrupt TMD organization. Pathogenic variants have not yet been identified with the cytoplasmic tail of SLC35A2, and previous reports suggest this region might be dispensable for its function (Sosicka, Jakimowicz, Olczak & Olczak, 2014).
Different topology prediction programs do not all agree. For example, four independent in silico tools suggest eight TMD and cytoplasmic loop 2 linking TMD2 and TMD3 is ~81aa (88–169), while in UNIPROT predicts ten TMD and cytoplasmic loop 2 with 8aa. (Supp. Figure S5). Although crystal structure for SLC35A2 has not been solved, the GDP-mannose transporter from Saccharomyces cerevisiae (vrg4) has, and clearly shows ten TMD (Parker & Newstead, 2017). Using this template, we modeled SLC35A2 3D using the PHYRE2 server (Kelley, Mezulis, Yates, Wass & Sternberg, 2015) producing ten TMD, with a slightly different distribution than that assigned by UNIPROT or other in silico prediction tools (Supp. Figure S5). Still, the majority of variants were localized to TMD (Fig. 1).
By combining the data from the UDP-galactose transport assay and cDNA allele ratio analysis allows an estimate of the activity of each SLC35A2 mutant. Based on these data, UDP-galactose transporter activity is reduced by ~70% in CDG-0187 when compared to the wild type protein, while CDG-0389 (~95%), CDG-0416 (~85%), CDG-0460 (~45%), CDG-0468 (~75%) and CDG-1039 (~40%) are also significantly reduced (Fig. 5B). However, this assumption does not consider the total amount of SLC35A2 protein based on western blot. For example, the level of UDP-galactose transporter protein in CDG-0389 fibroblasts is dramatically reduced compared to both control cells as well as other SLC35A2-CDG fibroblast lines.
The clinical and biochemical severity of females could vary due to XCI in different cell types. For instance, in CDG-0389, XCI from buccal and fibroblast were heavily skewed indicating “Non-random” inactivation while blood samples were not. Importantly, XCI results from cultured fibroblast support those seen using our allele ratio assay and western blot results.
While there are no available therapies for SLC35A2-CDG, a single case report shows an improved CDT profile in response to oral D-galactose (Dorre et al., 2015). However, it is unclear whether this response was due to D-galactose, because we know that SLC35A2-CDG CDT profiles change over time, (Ng et al., 2013). Controlled, long-term studies with clinically relevant measurements will be required to assess any benefits of oral D-galactose as a therapy for these individuals.
In conclusion, we present clinical, molecular and functional biochemical data on 30 individuals with SLC35A2-CDG to further expand the spectrum of this rare disorder and increases the number of known subjects to 62. We validated the UDP-Galactose transporter assay in all 5 available subject fibroblasts. Cells from the other 25 subjects were unavailable and must therefore be considered as likely pathogenic variants.
Supplementary Material
ACKNOWLEDGMENTS
We thank the families for their continued support. TMP was funded by the Cedars-Sinai Diana and Steve Marienhoff Fashion Industries Guild Endowed Fellowship in Pediatric Neuromuscular Diseases. We thank Jamie Smolin for her assistance.
FUNDING
This work is supported by The Rocket Fund, National Institutes of Health (NIH) grants R01DK099551 (to H.H.F) and personal contributions from Bill and Dinah Ruch and from Liberty Ross and Jimmy Iovine. National Science Center, 2016/21/B/NZ5/00144 (to M.O.)
The National Institute of Neurological Disorders and Stroke (NINDS) award number K08NS092898 and Jordan’s Guardian Angels (to G.M.). WBD supported by grants 5R01NS050375 and 1R01NS058721 from the NIH (NINDS).
Partially funded by the National Human Genome Research Institute Intramural Program.
Research reported in this manuscript was partially supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
UDN Award Numbers
Clinical sites
U01HG007709 (Baylor College of Medicine)
U01HG007672 (Duke University)
U01HG007690 (Harvard Teaching Hospitals)
U01HG007708 (Stanford Medicine)
U01HG007703 (University of California Los Angeles)
U01HG007674 (Vanderbilt University)
Coordinating Center
U01HG007530 (Harvard Medical School)
Sequencing Cores
U01HG007942 (Baylor College of Medicine)
U01HG007943 (HudsonAlpha Institute for Biotechnology)
Metabolomics Core
U01TR001395 (Battelle Pacific Northwest Laboratories)
Model Organisms Screening Center
U54NS093793 (Baylor College of Medicine)
CONFLICT OF INTEREST STATEMENT
The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from clinical genetic testing completed at Baylor Genetics Laboratories. Fernando Scaglia is involved in clinical trials supported by BioElectron, Reata Pharmaceuticals, and Stealth BioTherapeutics. Mary Willis declares views expressed herein are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, or the U.S. Government. Zöe Powis, Jesse M. Hunter, Sha Tang are employed by Ambry Genetics. Sharon F. Suchy, Jane Juusola, Dianalee McKnight, Maria Guillen Sacato and Hui Yang are employed by GeneDx, Inc., a wholly-owned subsidiary of OPKO Health, Inc. The other authors have no conflict.
REFERENCES
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, & Belmont JW (1992). Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet, 51(6), 1229–1239. [PMC free article] [PubMed] [Google Scholar]
- Bosch DG, Boonstra FN, de Leeuw N, Pfundt R, Nillesen WM, de Ligt J, … de Vries BB (2016). Novel genetic causes for cerebral visual impairment. Eur J Hum Genet, 24(5), 660–665. doi: 10.1038/ejhg.2015.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandli AW, Hansson GC, Rodriguez-Boulan E, & Simons K (1988). A polarized epithelial cell mutant deficient in translocation of UDP-galactose into the Golgi complex. J Biol Chem, 263(31), 16283–16290. [PubMed] [Google Scholar]
- Brockhausen I, & Stanley P (2015). O-GalNAc Glycans In rd, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, & Seeberger PH (Eds.), Essentials of Glycobiology (pp. 113–123). Cold Spring; Harbor (NY). [Google Scholar]
- Bruneel A, Cholet S, Drouin-Garraud V, Jacquemont ML, Cano A, Megarbane A, … Fenaille F (2018). Complementarity of electrophoretic, mass spectrometric, and gene sequencing techniques for the diagnosis and characterization of congenital disorders of glycosylation. Electrophoresis. doi: 10.1002/elps.201800021 [DOI] [PubMed] [Google Scholar]
- Chen J, Lu L, Shi S, & Stanley P (2006). Expression of Notch signaling pathway genes in mouse embryos lacking beta4galactosyltransferase-1. Gene Expr Patterns, 6(4), 376–382. doi: 10.1016/j.modgep.2005.09.009 [DOI] [PubMed] [Google Scholar]
- de Lonlay P, Seta N, Barrot S, Chabrol B, Drouin V, Gabriel BM, … Cormier-Daire V (2001). A broad spectrum of clinical presentations in congenital disorders of glycosylation I: a series of 26 cases. J Med Genet, 38(1), 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher SL, Nuwayhid N, Stanley P, Briles EI, & Hirschberg CB (1984). Translocation across Golgi vesicle membranes: a CHO glycosylation mutant deficient in CMP-sialic acid transport. Cell, 39(2 Pt 1), 295–299. [DOI] [PubMed] [Google Scholar]
- Dorre K, Olczak M, Wada Y, Sosicka P, Gruneberg M, Reunert J, … Marquardt T (2015). A new case of UDP-galactose transporter deficiency (SLC35A2-CDG): molecular basis, clinical phenotype, and therapeutic approach. J Inherit Metab Dis, 38(5), 931–940. doi: 10.1007/s10545-015-9828-6 [DOI] [PubMed] [Google Scholar]
- Etchison JR, Srikrishna G, & Freeze HH (1995). A novel method to co-localize glycosaminoglycan-core oligosaccharide glycosyltransferases in rat liver Golgi. Co-localization of galactosyltransferase I with a sialyltransferase. J Biol Chem, 270(2), 756–764. [DOI] [PubMed] [Google Scholar]
- Etchison JR, & Freeze HH (1996). A new approach to mapping co-localization of multiple glycosyl transferases in functional Golgi preparations. Glycobiology, 6(2), 177–189. [DOI] [PubMed] [Google Scholar]
- Euro, E.-R. E. S. C., Epilepsy Phenome/Genome, P., & Epi, K. C. (2014). De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet, 95(4), 360–370. doi: 10.1016/j.ajhg.2014.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira CR, Altassan R, Marques-Da-Silva D, Francisco R, Jaeken J, & Morava E (2018). Recognizable phenotypes in CDG. J Inherit Metab Dis, 41(3), 541–553. doi: 10.1007/s10545-018-0156-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze HH (2006). Genetic defects in the human glycome. Nat Rev Genet, 7(7), 537–551. doi: 10.1038/nrg1894 [DOI] [PubMed] [Google Scholar]
- Freeze HH, Eklund EA, Ng BG, & Patterson MC (2012). Neurology of inherited glycosylation disorders. Lancet Neurol, 11(5), 453–466. doi: 10.1016/S1474-4422(12)70040-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeze HH, Eklund EA, Ng BG, & Patterson MC (2015). Neurological aspects of human glycosylation disorders. Annu Rev Neurosci, 38, 105–125. doi: 10.1146/annurev-neuro-071714-034019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galupa R, & Heard E (2018). X-Chromosome Inactivation: A Crossroads Between Chromosome Architecture and Gene Regulation. Annu Rev Genet, 52, 535–566. doi: 10.1146/annurev-genet-120116-024611 [DOI] [PubMed] [Google Scholar]
- Grunewald S (2009). The clinical spectrum of phosphomannomutase 2 deficiency (CDG-Ia). Biochim Biophys Acta, 1792(9), 827–834. doi: 10.1016/j.bbadis.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Guillen E, Abeijon C, & Hirschberg CB (1998). Mammalian Golgi apparatus UDP-N-acetylglucosamine transporter: molecular cloning by phenotypic correction of a yeast mutant. Proc Natl Acad Sci U S A, 95(14), 7888–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Yamauchi M, Takahashi E, Hoshino M, Aoki K, Ayusawa D, & Kawakita M (1993). The UDP-galactose translocator gene is mapped to band Xp11.23-p11.22 containing the Wiskott-Aldrich syndrome locus. Somat Cell Mol Genet, 19(6), 571–575. [DOI] [PubMed] [Google Scholar]
- Hayes BK, Freeze HH, & Varki A (1993). Biosynthesis of oligosaccharides in intact Golgi preparations from rat liver. Analysis of N-linked glycans labeled by UDP-[6–3H]N-acetylglucosamine. J Biol Chem, 268(22), 16139–16154. [PubMed] [Google Scholar]
- Ichikawa M, Scott DA, Losfeld ME, & Freeze HH (2014). The metabolic origins of mannose in glycoproteins. J Biol Chem, 289(10), 6751–6761. doi: 10.1074/jbc.M113.544064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida N, Miura N, Yoshioka S, & Kawakita M (1996). Molecular cloning and characterization of a novel isoform of the human UDP-galactose transporter, and of related complementary DNAs belonging to the nucleotide-sugar transporter gene family. J Biochem, 120(6), 1074–1078. [DOI] [PubMed] [Google Scholar]
- Ishida N, Yoshioka S, Iida M, Sudo K, Miura N, Aoki K, & Kawakita M (1999). Indispensability of transmembrane domains of Golgi UDP-galactose transporter as revealed by analysis of genetic defects in UDP-galactose transporter-deficient murine had-1 mutant cell lines and construction of deletion mutants. J Biochem, 126(6), 1107–1117. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, & Sternberg MJ (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc, 10(6), 845–858. doi: 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Miura Y, Etchison JR, & Freeze HH (2001). Intact Golgi synthesize complex branched O-linked chains on glycoside primers: evidence for the functional continuity of seven glycosyltransferases and three sugar nucleotide transporters. Glycoconj J, 18(8), 623–633. [DOI] [PubMed] [Google Scholar]
- Kimizu T, Takahashi Y, Oboshi T, Horino A, Koike T, Yoshitomi S, … Imai K (2017). A case of early onset epileptic encephalopathy with de novo mutation in SLC35A2: Clinical features and treatment for epilepsy. Brain Dev, 39(3), 256–260. doi: 10.1016/j.braindev.2016.09.009 [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, & Shendure J (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet, 46(3), 310–315. doi: 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodera H, Nakamura K, Osaka H, Maegaki Y, Haginoya K, Mizumoto S, … Saitsu H (2013). De novo mutations in SLC35A2 encoding a UDP-galactose transporter cause early-onset epileptic encephalopathy. Hum Mutat, 34(12), 1708–1714. doi: 10.1002/humu.22446 [DOI] [PubMed] [Google Scholar]
- Lacey JM, Bergen HR, Magera MJ, Naylor S, & O’Brien JF (2001). Rapid determination of transferrin isoforms by immunoaffinity liquid chromatography and electrospray mass spectrometry. Clin Chem, 47(3), 513–518. [PubMed] [Google Scholar]
- Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P, … Gilissen C (2016). Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci, 19(9), 1194–1196. doi: 10.1038/nn.4352 [DOI] [PubMed] [Google Scholar]
- Lindahl U, Couchman J, Kimata K, & Esko JD (2015). Proteoglycans and Sulfated Glycosaminoglycans In rd, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, & Seeberger PH (Eds.), Essentials of Glycobiology (pp. 207–221). Cold Spring; Harbor (NY). [Google Scholar]
- Maszczak-Seneczko D, Olczak T, Wunderlich L, & Olczak M (2011). Comparative analysis of involvement of UGT1 and UGT2 splice variants of UDP-galactose transporter in glycosylation of macromolecules in MDCK and CHO cell lines. Glycoconj J, 28(7), 481–492. doi: 10.1007/s10719-011-9348-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maszczak-Seneczko D, Olczak T, Jakimowicz P, & Olczak M (2011). Overexpression of UDP-GlcNAc transporter partially corrects galactosylation defect caused by UDP-Gal transporter mutation. FEBS Lett, 585(19), 3090–3094. doi: 10.1016/j.febslet.2011.08.038 [DOI] [PubMed] [Google Scholar]
- Miura N, Ishida N, Hoshino M, Yamauchi M, Hara T, Ayusawa D, & Kawakita M (1996). Human UDP-galactose translocator: molecular cloning of a complementary DNA that complements the genetic defect of a mutant cell line deficient in UDP-galactose translocator. J Biochem, 120(2), 236–241. [DOI] [PubMed] [Google Scholar]
- Ng BG, Buckingham KJ, Raymond K, Kircher M, Turner EH, He M, … Freeze HH (2013). Mosaicism of the UDP-galactose transporter SLC35A2 causes a congenital disorder of glycosylation. Am J Hum Genet, 92(4), 632–636. doi: 10.1016/j.ajhg.2013.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng BG, & Freeze HH (2018). Perspectives on Glycosylation and Its Congenital Disorders. Trends Genet, 34(6), 466–476. doi: 10.1016/j.tig.2018.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelmann S, Stanley P, & Gerardy-Schahn R (2001). Point mutations identified in Lec8 Chinese hamster ovary glycosylation mutants that inactivate both the UDP-galactose and CMP-sialic acid transporters. J Biol Chem, 276(28), 26291–26300. doi: 10.1074/jbc.M011124200 [DOI] [PubMed] [Google Scholar]
- Olczak M, & Guillen E (2006). Characterization of a mutation and an alternative splicing of UDP-galactose transporter in MDCK-RCAr cell line. Biochim Biophys Acta, 1763(1), 82–92. doi: 10.1016/j.bbamcr.2005.12.006 [DOI] [PubMed] [Google Scholar]
- Parker JL, & Newstead S (2017). Structural basis of nucleotide sugar transport across the Golgi membrane. Nature, 551(7681), 521–524. doi: 10.1038/nature24464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portner A, Etchison JR, Sampath D, & Freeze HH (1996). Human melanoma and Chinese hamster ovary cells galactosylate n-alkyl-beta-glucosides using UDP gal:GlcNAc beta 1,4 galactosyltransferase. Glycobiology, 6(1), 7–13. [DOI] [PubMed] [Google Scholar]
- Sarkar AK, Fritz TA, Taylor WH, & Esko JD (1995). Disaccharide uptake and priming in animal cells: inhibition of sialyl Lewis X by acetylated Gal beta 1-->4GlcNAc beta-O-naphthalenemethanol. Proc Natl Acad Sci U S A, 92(8), 3323–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Rostand KS, Jain RK, Matta KL, & Esko JD (1997). Fucosylation of disaccharide precursors of sialyl LewisX inhibit selectin-mediated cell adhesion. J Biol Chem, 272(41), 25608–25616. [DOI] [PubMed] [Google Scholar]
- Schnaar RL, & Kinoshita T (2015). Glycosphingolipids In rd, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, & Seeberger PH (Eds.), Essentials of Glycobiology (pp. 125–135). Cold Spring; Harbor (NY). [PubMed] [Google Scholar]
- Sim NS, Seo Y, Lim JS, Kim WK, Son H, Kim HD, … Lee JH (2018). Brain somatic mutations in SLC35A2 cause intractable epilepsy with aberrant N-glycosylation. Neurol Genet, 4(6), e294. doi: 10.1212/NXG.0000000000000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosicka P, Jakimowicz P, Olczak T, & Olczak M (2014). Short N-terminal region of UDP-galactose transporter (SLC35A2) is crucial for galactosylation of N-glycans. Biochem Biophys Res Commun, 454(4), 486–492. doi: 10.1016/j.bbrc.2014.10.098 [DOI] [PubMed] [Google Scholar]
- Sosicka P, Maszczak-Seneczko D, Bazan B, Shauchuk Y, Kaczmarek B, & Olczak M (2017). An insight into the orphan nucleotide sugar transporter SLC35A4. Biochim Biophys Acta Mol Cell Res, 1864(5), 825–838. doi: 10.1016/j.bbamcr.2017.02.002 [DOI] [PubMed] [Google Scholar]
- Sosicka P, Bazan B, Maszczak-Seneczko D, Shauchuk Y, Olczak T, & Olczak M (2019) SLC35A5 Protein - A Golgi Complex Member with Putative Nucleotide Sugar Transport Activity. Int J Mol Sci, 20(2), 276 doi.org/10.3390/ijms20020276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P (1981). Selection of specific wheat germ agglutinin-resistant (WgaR) phenotypes from Chinese hamster ovary cell populations containing numerous lecR genotypes. Mol Cell Biol, 1(8), 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley P, Taniguchi N, & Aebi M (2015). N-Glycans In rd, Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, Schnaar RL, & Seeberger PH (Eds.), Essentials of Glycobiology (pp. 99–111). Cold Spring; Harbor (NY). [PubMed] [Google Scholar]
- Toma L, Pinhal MA, Dietrich CP, Nader HB, & Hirschberg CB (1996). Transport of UDP-galactose into the Golgi lumen regulates the biosynthesis of proteoglycans. J Biol Chem, 271(7), 3897–3901. [DOI] [PubMed] [Google Scholar]
- Westenfield K, Sarafoglou K, Speltz LC, Pierpont EI, Steyermark J, Nascene D, … Pierpont ME (2018). Mosaicism of the UDP-Galactose transporter SLC35A2 in a female causing a congenital disorder of glycosylation: a case report. BMC Med Genet, 19(1), 100. doi: 10.1186/s12881-018-0617-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winawer MR, Griffin NG, Samanamud J, Baugh EH, Rathakrishnan D, Ramalingam S, … Heinzen EL (2018). Somatic SLC35A2 variants in the brain are associated with intractable neocortical epilepsy. Ann Neurol, 83(6), 1133–1146. doi: 10.1002/ana.25243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Zhang W, Li X, Jiang R, Harper T, Liu R, … He M (2013). Serum N-glycan and O-glycan analysis by mass spectrometry for diagnosis of congenital disorders of glycosylation. Anal Biochem, 442(2), 178–185. doi: 10.1016/j.ab.2013.07.037 [DOI] [PubMed] [Google Scholar]
- Yates TM, Suri M, Desurkar A, Lesca G, Wallgren-Pettersson C, Hammer TB, … Balasubramanian M (2018). SLC35A2-related congenital disorder of glycosylation: Defining the phenotype. Eur J Paediatr Neurol. doi: 10.1016/j.ejpn.2018.08.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.