Figure 2. Detection of Epitope-Tagged 3′ UTR Translation Products Results from 80S Reinitiation in the hcr1Δ Strain.
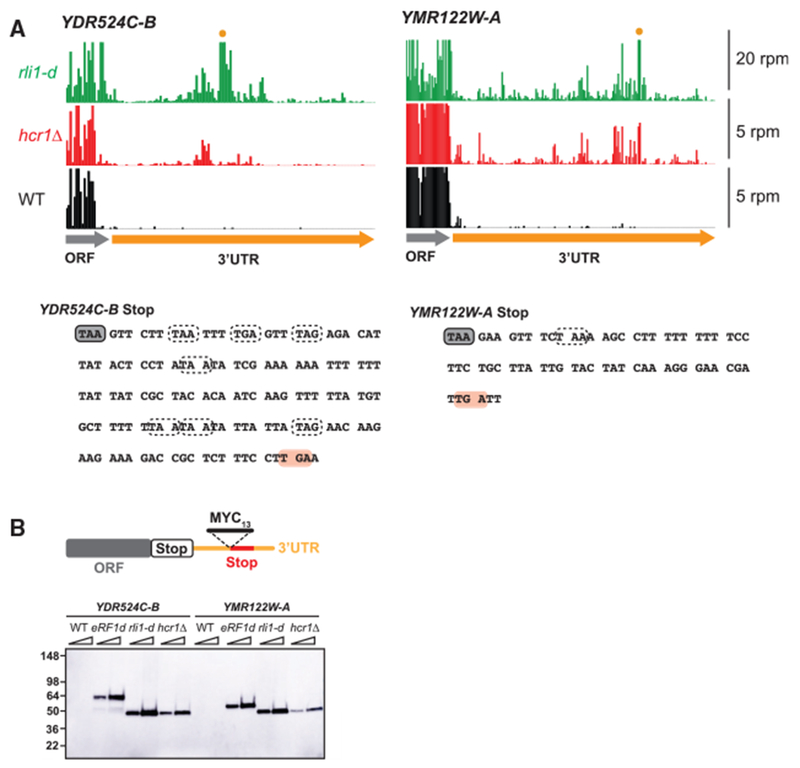
(A) Ribosome footprint profiles of the 3′ UTRs of genes YDR524C-B and YMR122W-A showing a similar pattern of footprint density between hcr1Δ and rli1-d cells (data from pooled replicates). The scale is zoomed in so reads in the ORF region are clipped. Two cases in which reads in the 3′ UTR for rli1-d exceed the scale because of a strong stop codon peaks are marked (orange dot). Note that reads from WT and hcr1Δ cells are scaled by 4x for better comparison. Sequences for each gene are shown below with stop codons in all frames indicated with dotted lines, except the stop codon corresponding to the schematic in (B) that the MYC13 tag is inserted in front of, which is indicated in pink.
(B) MYC13-tagged reporters that had previously been used to detect 80S reinitiation in the 3′ UTR of Rli1-depleted cells (Young et al., 2015) were inserted into the chromosome of hcr1Δ cells.
WT, eRF1d, rli1-d, and hcr1Δ cells carrying the MYC13- tagged reporters were grown to log-phase before reparation of whole cell extracts (WCEs). WCEs were prepared from five optical densities (ODs) of cells by trichloroacetic acid (TCA) extraction. WCEs were subjected to western analysis using an antibody against c-Myc. Two different amounts of protein were loaded for each sample (1 × and 2 ×). Migration of molecular weight (MW) standards (kDa) is indicated on the left. The 3′ UTR polypeptides in the hcr1Δ strain are of a similar size to those observed in the rli1-d strain and run at the expected weight (Young et al., 2015) for the tag plus a small amount of translated 3′ UTR sequence. In contrast, products from the eRF1-depleted strain run at a heavier weight because of inclusion of the entire protein encoded by the main ORF in the observed product.
