Figure 4. Overexpression of RLI1 Suppresses the Recycling Defect Observed in the hcr1Δ Strain.
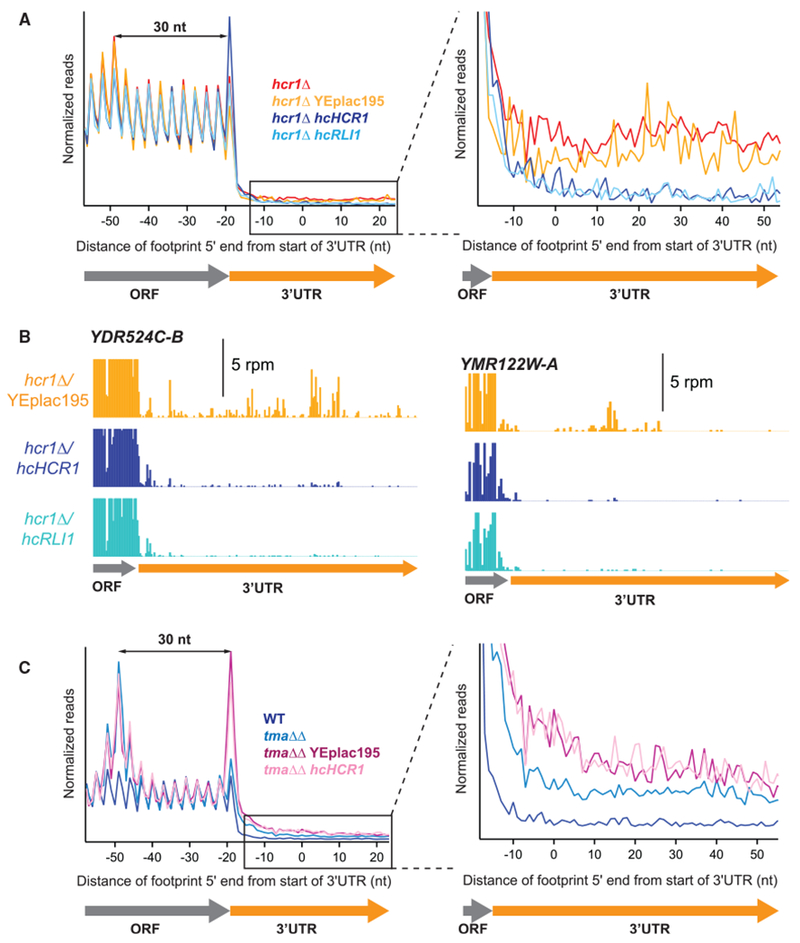
(A) Normalized average ribosome footprint occupancy (equally weighted by reads within the ORF) for all genes aligned at their stop codons. Footprints plotted by 5′ ends. (Inset) Magnified view of (A), showing complementation and suppression of 3′ UTR ribosome occupancy in the hcr1Δ hcHCR1 and hcr1Δ hcRLI1 strains, respectively.
(B) Individual examples of the elimination of 3′ UTR reads shown in the average in (A). Ribosome footprint profiles for the genes YDR524C-B and YMR122W-A showing loss of 3′ UTR footprint density in the hcr1Δ hcHCR1 and hcr1Δ hcRLI1 strains relative to hcr1Δ with an empty vector.
(C) Normalized average ribosome footprint occupancy as for (A) but showing the effect of overexpression of HCR1 on tma64Δ/tma20Δ. (Inset) Magnified view of (C), showing that 3′ UTR ribosome occupancy in the tma64Δ/tma20Δ strain is not suppressed by hcHCR1.
See also Figure S3.
