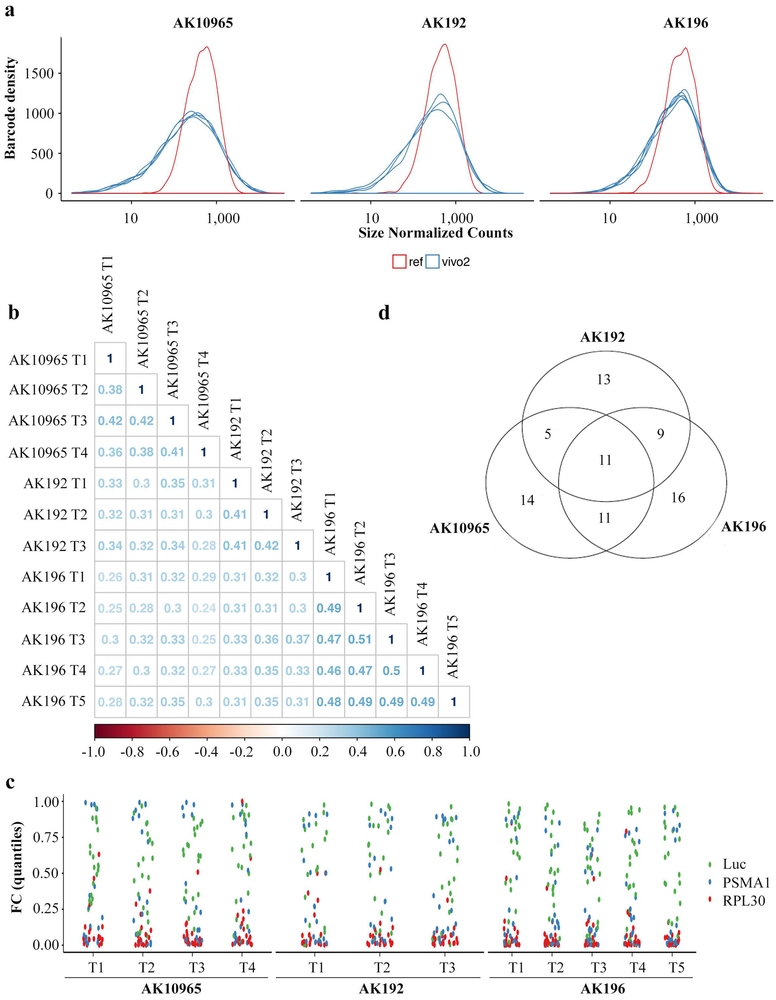
Extended Data Figure 2. Functional surfaceome analysis identified SDC1 as a KRAS*-dependent surface candidate.
a, Normalized counts showing the distribution of reference and tumor barcodes to establish library coverage in vivo in three independent iKras p53L/+ tumor cells cultures, which show similar results. In vivo screens were conducted in 3-5 mice for for each cell culture.
b, Correlation matrix among replicates of orthotopic xenograft-derived AK192, AK196, and AK10965 tumors screened with the surfacome-targeting shRNA library (barcode level fold-change comparison, Pearson correlation coefficient).
c, Positive (Psma, Rpl30) and negative (Luc) controls were plotted applying the RSP/LogP score.
d, Venn diagram shows the number of hits identified from three independent iKras p53L/+ tumor cell cultures.