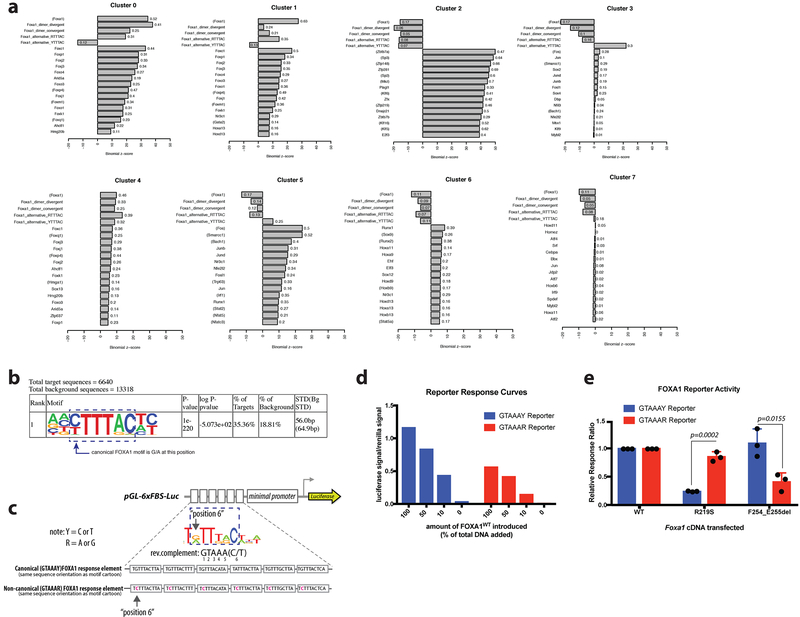
Extended Data Figure 7. Motif analysis of ATAC-sequencing and modification of FOXA1 reporter assay for evaluation of non-canonical FOXA1 motif.
(a) FIMO motif analysis of ATAC-seq clusters. Summary of motif enrichments/depletion results for each cluster relative to the background of all differentially accessible peaks, as reported by binomial Z-score. The top 15 enriched database motifs for expressed transcription factors are shown for each cluster. In addition, enrichment/depletion results for four additional FOXA1-related motifs are shown: convergent and divergent dimer motifs, and altered FOXA1 core binding motifs with either G/A or C/T at position 6. Transcription factors in parentheses represent motifs inferred from other species. Complete lists can be found in Supplementary Tables 3-10. (b) Top motif identified de novo using HOMER on ATAC-seq cluster 3 (R219S-specific) with motif core indicated, and variation from canonical FOXA1 motif depicted. p-values derived from one-sided binomial test. (c) Schematic of reporter design. Canonical response element reporter is same reporter used in Fig. 2, with various iterations of the canonical FOXA1 motif in tandem. Non-canonical motif has substitutions at position 6, indicated in pink, to reflect the newly identified motif enriched in cluster 3 of ATAC-seq. Note: the orientation of the upper motif cartoon and the sequence in the reporter schematic are the reverse complement of the motif identified by HOMER (GTAAAR). Modified base noted in position 6. (d) Dose response curve for both FOXA1 luciferase reporters’ activity in response to increased amounts of Foxa1WT cDNA introduced into the system. Data shown is one representative biological replicate of 3 carried out, all showing same trends, but absolute luciferase/renilla ratios vary from experiment to experiment. (e) Results of reporter assays expressed as a relative response ratio, normalized to level of FOXA1WT activity for a given reporter. Data from 3 biological replicates, bars indicate mean +/− standard deviation. p-values derived using unpaired, two-tailed Student’s T-test.