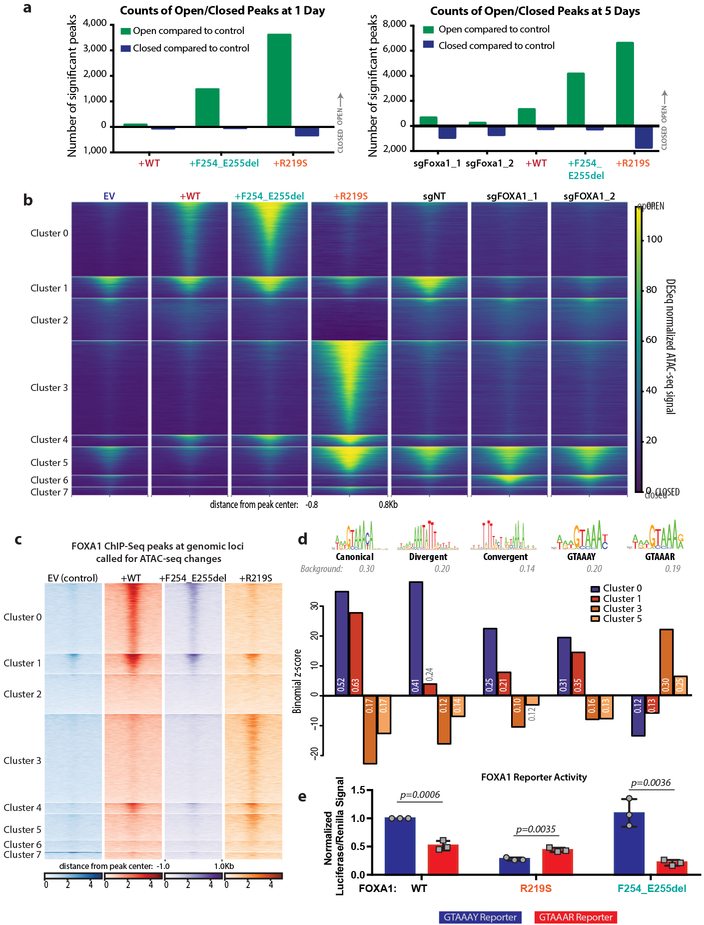
Fig. 4. FOXA1 mutations cause dramatic shifts in the chromatin landscape.
(a) Number of significant peaks open or closed (log2FC >2 for open, <−2 for closed peaks) after dox treatment for pCW-FOXA1wild-type or mutant organoids relative to EV. Right panel includes counts for FOXA1 CRISPR organoids 5 days after trypsinization relative to sgNT. Data from 3 biological replicates, with FDR <0.05 using two-sided Wald test, with Benjamini-Hochberg FDR correction for multiple observations. (b) ATAC-seq peak heat maps comparing organoids with (sgFOXA1_1, sgFOXA1_2) or without (sgNT) CRISPR deletion of Foxa1 or expression of WT or mutant FOXA1 after 5 days of dox treatment, with eight clusters defined by hierarchical clustering. (c) FOXA1 ChIP-seq signal at genomic loci matching ATAC-seq clusters defined in panel b shows a similar pattern of peaks correlating FOXA1 binding with open chromatin. Data from two biological replicates. (d) Enrichment or depletion of FOXA1 motif variants in clusters that gain accessibility in +F254_E255del or +R219S organoids, including the canonical motif, divergent and convergent dimer motifs, and altered versions of the FOXA1 motif (GTAAAY, similar to canonical and GTAAAR, non-canonical), expressed as a binomial Z-score computed from the number of cluster peaks with >1 motif occurrence relative to background occurrence in all heatmap peaks. Occurrence within a given cluster is reported within the bar graph. Positive scores indicate enrichment; negative scores indicate depletion. (e) Luciferase reporter assay depicting activity of FOXA1 variants on GTAAAY (blue) or GTAAAR (red) DNA templates. Luciferase/Renilla signal normalized to signal from FOXA1WT on GTAAAY reporter. Data from 3 biological replicates represented as mean +/− standard deviation. p-values determined using unpaired, two-tailed Student’s T-test. No significant difference between activity of WT and R219S on the GTAAAR reporter (p= 0.2314). F254_E255del has significantly less activity on GTAAAR than either WT (p= 0.0059) or R219S (p= 0.0033).