Figure 1.
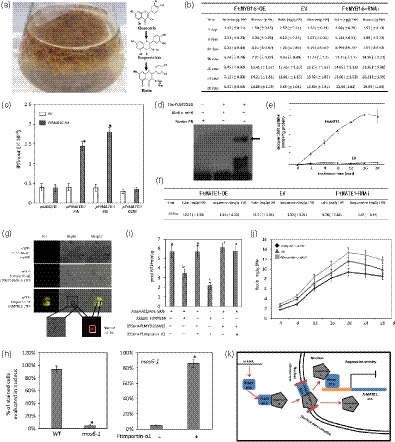
FtMYB16 regulates FtMATE1 for rutin biosynthesis. (a) Rutin biosynthesis pathway and hairy root cultures of F. tataricum. (b) Rutin content and fresh weight in different genotypes of 4‐, 8‐, 12‐, 16‐, 20‐, 24‐ and 28‐day‐old F. tataricum hairy roots. Values are means ± SD of three biological repeats of each independent transgenic line. The transgenic hairy root lines generated from transformations with strains A. rhizogenes A4 harbouring T‐DNA contain no open reading frame (empty vector, EV; Lines 1, 3, 4), FtMYB16‐OE (Lines 1, 2, 4) and FtMYB16‐RNAi (Lines 1, 4, 6). (c) Direct binding of FtMYB16 with the promoter of FtMATE1 by ChIP assays. ChIP assays were conducted by RT‐PCR after normalizing with the input DNA. The fragment of FtMATE1 coding sequence and the reference gene UBQ10 promoter were used as a negative control. Significant differences between values are indicated with asterisks as tested by Student's t‐test (P < 0.05). (d) EMSA of a probe or mutant probe FtMATE1 promoter fragment B and its mutant with His‐FtMYB16 purified from Escherichia coli BL21 (DE3). The arrow indicates the protein probe complex. (e) Time‐dependent uptake into vesicles from yeast cells transformed with FtMATE1 or empty vector (EV) to 100 μm isoquercitrin. (f) Rutin and isoquercitrin contents in different genotypes of 20‐day‐old F. tataricum hairy roots. The transgenic hairy root lines generated from transformations with strains A. rhizogenes A4 harbouring T‐DNA contain no open reading frame (empty vector, EV; Lines 1, 2, 3), FtMATE1‐OE (Lines 3, 6, 9) and FtMATE1‐RNAi (Lines 2, 4, 5). Values are means ± SD of three biological repeats of each independent transgenic line. Letters indicate statistically significant differences compared with each other (P < 0.05, post hoc Tukey's HSD test). (g) FtMYB16 interacts with Ftimportin‐α1 in planta by BiFC assays. YFP fluorescence images alone or merged with bright‐field images of Arabidopsis cell suspension protoplasts co‐transfected with constructs encoding the indicated fusion proteins with YFP at the C‐terminus or the N‐terminus, RFP nuclear marker as a nuclear‐localized analysis. Scale bar = 10 μm. (h) Quantification of YFP‐FtMYB16 nuclear localization or with Ftimportin‐α1 in WT and mos6‐1 mutant plant mesophyll protoplasts. At least 200 mesophyll protoplasts from each transformation were counted. Error bars indicate the ± SD from three independent biological replicates. Asterisks indicate statistically significant differences compared with each other (P < 0.05, Student's t‐test). (i) Arabidopsis cell suspension protoplasts were cotransformed with 2 μg reporter plasmid of FtMATE1 pro1‐GUS and 2 μg of effector plasmids of 35S:: FtMYB16, 35S:: FtMYB16 ΔNLS and 35S:: Ftimportin‐α1. Values represent means ± SE of triplicate experiments. Letters indicate statistically significant differences compared with each other (P < 0.05, post hoc Tukey's HSD test). (j) Rutin contents in different genotypes of 4‐, 8‐, 12‐, 16‐, 20‐, 24‐ and 28‐day‐old F. tataricum hairy roots. The transgenic hairy root lines generated from transformations with strains A. rhizogenes A4 harbouring T‐DNA contain no open reading frame (empty vector, EV; Lines 2, 4, 5), Ftimportin‐α1‐OE (Lines 1, 3, 6) and Ftimportin‐α1‐RNAi (Lines 1, 3, 9). Values are means ± SD of three biological repeats of each independent transgenic line. (k) A model of the role of Ftimportin‐α1 in the regulation of the FtMYB16 nucleo‐cytoplasmic trafficking in mediating rutin metabolism in Fagopyrum. The cytoplasmic cargo protein FtMYB16 binds to importin‐α Ftimportin‐α1; then, the complex FtMYB16/importin‐α translocates FtMYB16 into the nucleus, and this nucleo‐cytoplasmic trafficking signalling pathway depends on their protein interaction domain, such as NLS motif. FtMYB16 not only represses some key enzyme genes, such as FtPAL, in rutin biosynthesis, but also represses the gene expression of MATE transporter FtMATE1 which transports isoquercitrin for rutin metabolism. Additionally, Ftimportin‐α1 functions as a corepressor on the transcriptional activity of FtMYB16 on its target genes.
