Figure 4.
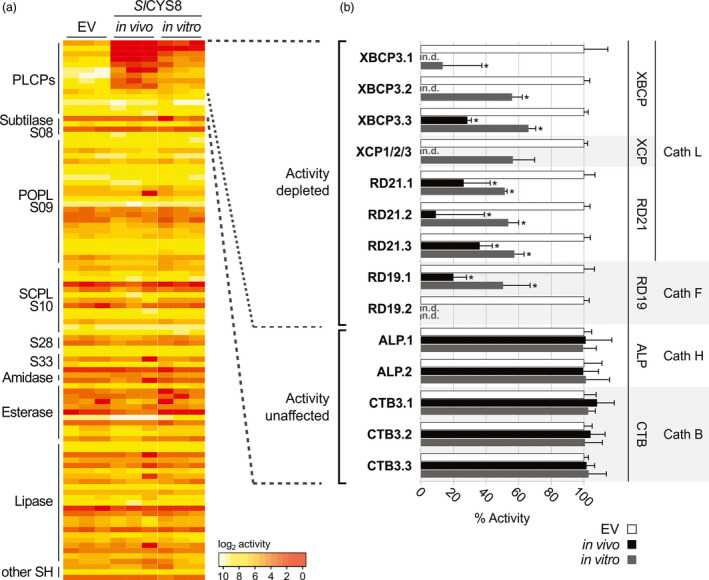
Activity‐based proteomics reveals nine endogenous PLCPs that are inhibited by Sl CYS8. Total leaf extracts were isolated from plants expressing Sl CYS8 (in vivo) or expressing an empty vector (EV), pre‐incubated with or without 280 nm of purified Sl CYS8 (in vitro). Proteins were labelled with DCG04 and FP‐biotin to label PLCPs and Ser hydrolases, respectively. Labelled proteins were purified and identified by MS. (a) Heatmap showing active enzymes that were significantly enriched (FDR < 0.05) compared to a no‐probe control, fold inhibition serves as a proxy for activity. Most PLCP activities were depleted upon Sl CYS8 expression, while no Ser hydrolase was inhibited. (b) Identified PLCPs were grouped by subfamilies and the activity was normalized to the EV control (100%). Bars are the means of three biological replicates ± SE. Star indicates significant differences from the EV control (Student's t test, P < 0.05). PLCP subfamilies: Xylem and bark Cys proteases (XBCPs), Xylem‐specific Cys proteases (XCPs), Resistant‐to‐Desiccation‐21 proteases (RD21s), Resistant‐to‐Desiccation‐19 proteases (RD19s), Aleurain‐Like proteases (ALPs) and Cathepsin‐B‐like proteases (CTBs). n.d.; not detected. MS dataset is presented in Table S1.
