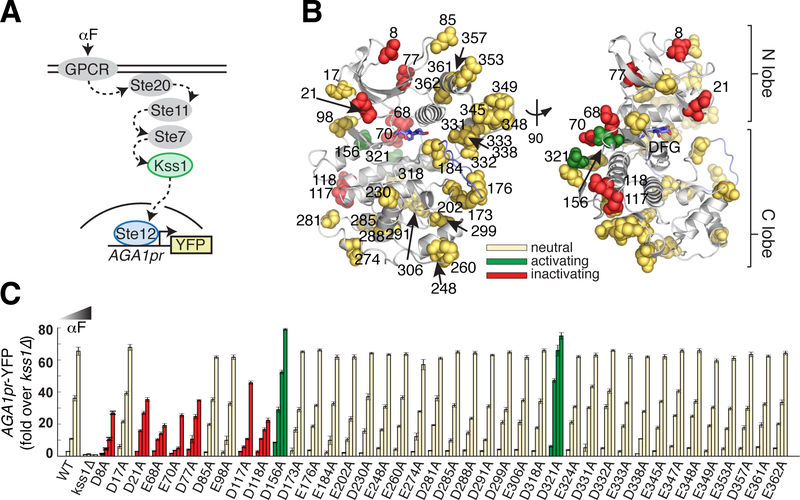
Figure 2. Alanine scan of acidic residues on the solvent accessible surface of yeast MAPK Kss1.
A. Schematic of the Kss1-dependent yeast pheromone pathway. The alpha factor (αF) mating pheromone binds to a G-protein coupled receptor (GPCR), leading to activation of a signaling cascade culminating at the MAPK Kss1. Kss1 then activates the Ste12 transcription factor to induce the mating transcriptional program, which can be monitored by fusing the promoter of the target gene AGA1 to a YFP reporter.
B. Ribbon diagram of a Kss1 homology model (30) with the 40 solvent accessible Asp/Glu residues shown as spheres. The DFG motif and activation loop are indicated in light blue. All 40 positions were mutated individually to alanine to remove negative charge.
C. The 40 resulting yeast strains along with wild type and kss1Δ controls were assayed for activation of the AGA1pr-YFP reporter by flow cytometry following treatment with 0, 0.01, 0.1 and 1 μM αF for 4 hours. Bars represent the average of the median YFP fluorescence from 3 biological replicates normalized to the untreated kss1Δ cells, and error bars are the standard deviation of the biological replicates. Mutations at red and green positions resulted in significantly reduced or increased YFP expression (P < 0.05, as scored by one-way ANOVA) in response to at least two doses of αF, respectively. Yellow positions indicate that the mutation had no effect in this assay. The color coding is identical in (B). Nine acidic positions on the solvent accessible surface are functionally coupled to kinase activity.