Figure 6.
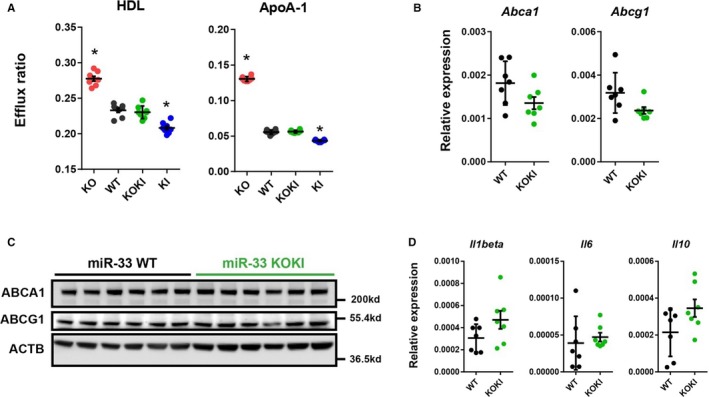
Similar characteristics of peritoneal macrophages obtained from microRNA (miR)‐33 wild‐type (WT) and miR‐33 knock out knock in (KOKI) mice. A, Cholesterol efflux to high‐density lipoprotein (HDL) or apolipoprotein A‐1 (ApoA‐1) from peritoneal macrophages from the 4 strains of mice. n=8 for each. B, Quantitative polymerase chain reaction (PCR) analysis for miR‐33 target genes in the peritoneal macrophages. n=7 for each. C, Representative images of Western blotting analysis for miR‐33 target genes in the peritoneal macrophages. n=6 for each. D, Quantitative PCR analysis for inflammatory markers in the peritoneal macrophages. n=7 for each. Horizontal bars in the dot plots indicate mean±SEM. ABCA indicates ATP‐binding cassette A; ABCG1, anti ATP binding cassete G1; ACTB, beta‐actin; KO, Knock out. *P<0.05 compared with WT.
