Figure 3.
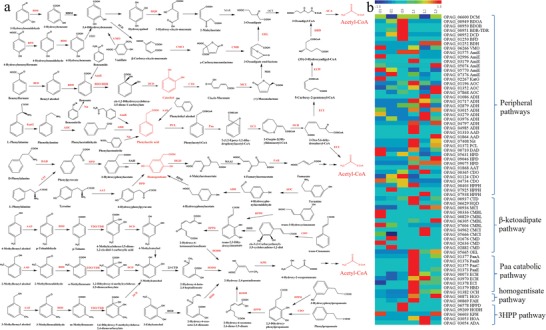
Regulation of catabolic pathways for lignin‐derived aromatic compounds in R. opacus PD630 as revealed by proteomics‐guided systems biology. a) The reconstructed catabolism pathways for lignin‐derived aromatic compounds based on proteomics analysis of R. opacus PD630. The proteins highlighted in red were detected by the proteomics, and those in black were found from genome but not detected by proteomics in this study. The three aromatic compounds highlighted in red color were the major intermediates for aromatic compound degradation. The metabolites were eventually funneled to acetyl‐CoA for degradation. b) The heatmap of relative expression abundance of differentially expressed proteins involved in aromatic compound catabolism. Each row of the heatmap represents one protein, and each column represented one condition. The data presented in the heatmap was the relative protein abundance normalized by each row (protein) among different conditions. The conditions included the three growth stages of R. opacus PD630 grown on 1% glucose (G) or lignin (L) as a sole carbon source: log phase (G1, L1), early stationary phase (G2, L2), and late stationary phase (G3, L3). The proteins presented in this figure were: AAD, aryl‐alcohol dehydrogenase; AAT, aspartate aminotransferase; ACA, acetyl‐CoA acyltransferase; ADA, acetaldehyde dehydrogenase; ADH, aldehyde dehydrogenase (NAD(P)+); AmiE, amidase; AOC, primary‐amine oxidase; BDH, benzaldehyde dehydrogenase (NAD); BDO, benzoate 1,2‐dioxygenase; BDOA, benzoate 1,2‐dioxygenase subunit alpha; BDOB, benzoate 1,2‐dioxygenase subunit beta; BDR, benzoate 1,2‐dioxygenase reductase component; BFD, benzoylformate decarboxylase; CDO, trans‐cinnamate dioxygenase; CMBL, carboxymethylenebutenolidase; CMCI,3‐carboxy‐cis,cis‐muconate cycloisomerase; CMD, 4‐carboxymuconolactone decarboxylase; CMDL, carbon‐monoxide dehydrogenase large subunit; CMDM, carbon‐monoxide dehydrogenase medium subunit; CTD, catechol 1,2‐dioxygenase; 23‐CTD, catechol 2,3‐dioxygenase; DAD, D‐amino‐acid dehydrogenase; DCD, dihydroxycyclohexadiene carboxylate dehydrogenase; DCM, 2,4‐dichlorophenol 6‐monooxygenase; ECH, enoyl‐CoA hydratase; ECI, 2‐(1,2‐epoxy‐1,2‐dihydrophenyl)acetyl‐CoA isomerase; FAH, fumarylacetoacetate hydrolase; HBD, 3‐hydroxybutyryl‐CoA dehydrogenase; HBM, 3‐hydroxybenzoate 4‐monooxygenase; HDH, 2‐hydroxy‐6‐oxo‐octa‐2,4‐dienoate hydrolase; HGO, homogentisate 1,2‐dioxygenase; HOA, 4‐hydroxy 2‐oxovalerate aldolase; HODH, 2‐hydroxy‐6‐oxonona‐2,4‐dienedioate hydrolase; HPD, 4‐hydroxyphenylpyruvate dioxygenase; HPM, 4‐hydroxyphenylacetate 1‐monooxygenase; HPPD, 2,3‐dihydroxyphenylpropionate 1,2‐dioxygenase; HPPH, 3‐(3‐hydroxy‐phenyl)propionate hydroxylase; HQD, Hydroxyquinol 1,2‐dioxygenase; KatG, catalase‐peroxidase; KPH, 2‐keto‐4‐pentenoate hydratase; MAAI, maleylacetoacetate isomerase; MAR, maleylacetate reductase; MCI, muconate cycloisomerase; Nit, nitrilase; OCH, oxepin‐CoA hydrolase; OCT, 3‐oxoadipate CoA‐transferase; OEL, 3‐oxoadipate enol‐lactonase; Paa, ring‐1,2‐phenylacetyl‐CoA epoxidase; PCD, protocatechuate 3,4‐dioxygenase; PCDA, protocatechuate 3,4‐dioxygenase, alpha subunit; PCDB, protocatechuate 3,4‐dioxygenase, beta subunit; PCL, phenylacetate‐CoA ligase; SAH, salicylate hydroxylase; TDO, toluate 1,2‐dioxygenase; TDOA, toluate 1,2‐dioxygenase subunit alpha; TDOB, toluate 1,2‐dioxygenase subunit beta; TDR, toluate 1,2‐dioxygenase reductase component; VMO, vanillate monooxygenase.
