Abstract
Precision medicine relies on validated biomarkers with which to better classify patients by their probable disease risk, prognosis and/or response to treatment. Although affordable ‘omics’-based technology has enabled faster identification of putative biomarkers, the validation of biomarkers is still stymied by low statistical power and poor reproducibility of results. This Review summarizes the successes and challenges of using different types of molecule as biomarkers, using lung cancer as a key illustrative example. Efforts at the national level of several countries to tie molecular measurement of samples to patient data via electronic medical records are the future of precision medicine research.
As announced by the US President Barack Obama during the 2015 State of the Union Address1, with further details provided by leaders at the US National Institutes of Health2,3, the Precision Medicine Initiative promises to improve human health by combining clinical data and biomarker measurements on a massive scale. The goal of these precision medicine efforts is to use multiple types of data to classify patients into precise groups that will benefit from a given treatment approach. Similar efforts have already begun in the United Kingdom4, in Denmark5 and in Germany5, where universal health care has made data collection easier. The term ‘precision medicine’ gained momentum with the publication of the 2011 US Institute of Medicine’s National Research Council report Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease6. This report summarizes a research pathway to redefine and unite the taxonomic systems by which the medical and scientific communities classify diseases. Patients with different biomarkers present with different risks of developing a disease, different disease prognoses or different responses to treatment; therefore, new biomarkers will be added to the current standards of phenotypic features (symptoms and histology) and medical history to revise the definition of a disease to include a new subtype (taxa) (FIG. 1). New standards of care will then be developed for these newly defined disease taxa. For example, epidermal growth factor receptor (EGFR)-positive non-small cell lung adenocarcinoma is currently its own taxon as opposed to being included in the general lung adenocarcinoma taxon and is treated with different chemotherapy from that of non-EGFR-driven adenocarcinomas7. The term precision medicine is new and arguably more accurate than its predecessor ‘personalized medicine’, but the overarching approach to developing improved biomarkers has not appreciably changed (BOX 1). Rather, the primary difference between the precision medicine research approach and traditional biomarker development is the magnitude of data collected and the speed at which data from different sources is (usually simultaneously) analysed. The Institute of Medicine’s report serves as a unified reference document that includes standard terminology that researchers of multiple disciplines can use when designing precision medicine studies.
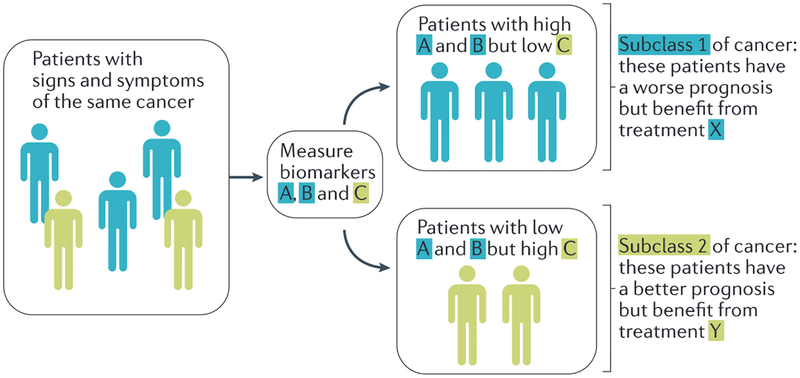
Figure 1|. Classifying patients into new, specific taxa.
Patients with the same signs and symptoms of cancer often have different outcomes. The precision medicine approach provides a research strategy to develop biomarkers that can be used to classify patients with the same cancer into finer taxa (subclass 1 versus subclass 2) by biomarkers that predict prognoses derived from the synthesis of large amounts of data to identify discriminating biomarkers. For example, patients in subclass 1 who have a worse prognosis (that is, have biomarkers that are associated with poor survival) may be given a more aggressive treatment (treatment X) versus those in subclass 1 who have a better prognosis (that is, have biomarkers that are associated with good outcome) and require a less aggressive therapy (treatment Y). Additionally, the converse may be true where individuals with a worse prognosis are provided less aggressive therapy if no benefit from aggressive treatment has been observed for this subclass.
Box 1|. A brief history of the term ‘precision medicine’.
Hippocrates gave the field of medicine the sound advice that “It is far more important to know what sort of person the disease has than what sort of disease the person has.” Thus, the notion that individuals presenting with the same signs and symptoms may require different treatment is not new. Precision medicine has a long list of predecessor terms with similar meaning, including personalized medicine, P4 (predictive, preventive, participatory and personalized) medicine, genomics medicine, predictive medicine and individualized medicine. Regardless of the name of the approach, the goal is to use molecular data in addition to more traditional clinical information (for example, symptoms, personal history and histology) to tailor medical care to provide the most benefit while minimizing risk. The application of precision medicine is anticipated to improve all areas of medicine, including predicting an individual’s risk of disease, disease prognosis and risk of side effects versus positive response to disease treatment approaches. Thus far, the greatest advances of precision medicine have been achieved in the prediction of response to a drug therapy using companion diagnostics (that is, biomarkers that can predict response to a specific drug treatment).
The precision with which a disease can be classified into subtypes (or taxa) rests heavily on the success of the research framework outlined in the Institute of Medicine’s report6 and on supportive mechanistic research. Specifically, this report proposes that an ‘Information Commons’ will serve as a giant reservoir of medical history, demographic, molecular measurement and disease outcome data (FIG. 2). The data mined and analysed from this Information Commons should then be integrated with other published biomedical literature on mechanisms of action to generate an educated Knowledge Network, which will identify biomarkers that classify patients by differential risk, diagnosis, response or outcome. Insights gained from the Knowledge Network will then be used to revise subtype definitions and standard of care for each disease subtype. This process of Information Commons-Knowledge Network-redefining taxon definitions is an iterative one that is ideally evaluated in real time to improve the precision with which patients can be diagnosed with a disease subtype. That is, the entire process is actively provided updated patient outcome data or relevant mechanistic data as soon as they are available to continually improve taxonomy, apply the new taxonomy and collect more data on patient outcomes using the new taxonomy. The real-time update of patient and biomarker data is a goal of the US Precision Medicine Initiative for its national cohort of 1,000,000 citizens; this Initiative will therefore be a true test of the limits of the precision medicine approach to define new disease taxonomy3.
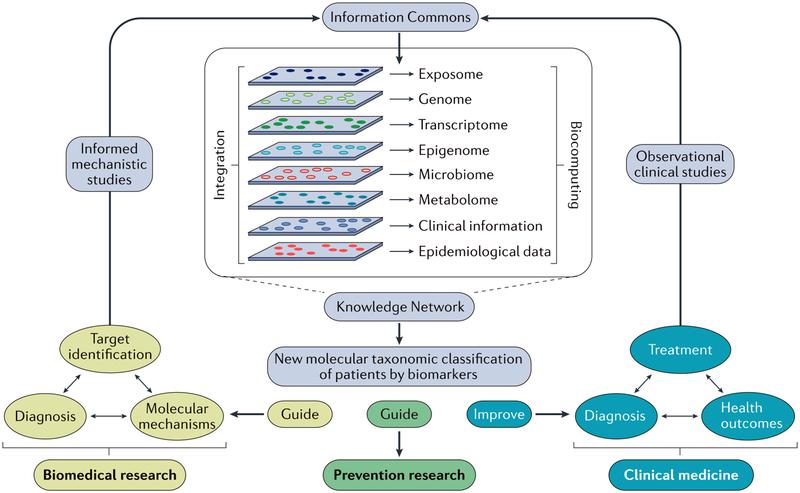
Figure 2 |. A precision medicine research strategy.
As outlined in the 2011 Institute of Medicines National Research Council report entitled Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease6, an Information Commons will be analysed to develop a Knowledge Network to inform research and medicine. The Information Commons will serve as a reservoir of data on a group of individuals from multiple sources (clinical data, demographic and epidemiological data, and multiple types of ‘omics’ data). Analyses of the Information Commons will result in the generation of a Knowledge Network that will specify clinical, demographic and ‘omics’ characteristics that predict disease risk, diagnosis, response and prognosis, thus allowing for the reclassification of individuals into subtypes (taxa). These new taxa will require further research and clinical follow-up to validate their existence and to determine the most suitable taxon-specific standards of care. Adapted with permission from REF. 6 by the National Academy of Sciences, Courtesy of the National Academies Press, Washington, DC, USA.
Biomarkers are the foundation of improving diagnostic precision. Biomarkers can be correlational (that is, only associated with disease) and/or functional (that is, they have an identified mechanism of action related to disease). Functional biomarkers can also be used as potential therapeutic targets. Biomarkers can be measured alone or in a group, often called a biomarker panel, to infer risk, diagnosis, prognosis and therapeutic response. DNA, RNA, proteins, metabolites, host cells and microorganisms can all function as biomarkers, and are now often measured using ‘omics’ methodology. Lower throughput, tissue visualization-and imaging-based biomarkers are also commonly used due to the availability of formalin-fixed, paraffin-embedded clinical specimens. Biomarkers can be measured in a variety of biological material (for example, blood, organ tissue, stool, saliva and urine). Analogous to the phases of drug development, Pepe et al.8 outlined a set of five phases to serve as an early guide for researchers aiming to bring biomarkers to the clinic. These five phases are as follows: preclinical, exploratory studies; clinical assay development and validation studies; longitudinal, retrospective studies; prospective, screening studies; and studies to determine whether the biomarker reduces morbidity or mortality in the population. The vast amount of data generated by precision medicine research is likely to affect each of these phases.
The development of biomarkers is directed by regulatory guidelines. In this regard, the US Food and Drug Administration (FDA) is recognized as a leader in the regulation of precision medicine because it was the first to set regulatory guidelines in this field9. For example, the FDA provides a guided pathway for the development of precision medicine biomarkers that are paired with a companion therapeutic agent. These paired biomarkers are termed companion diagnostics10. The European Union’s European Medicines Agency (EMA)11,12 and Japans Pharmaceutical and Medical Device Agency (PMDA)13 have also released guidance for companion diagnostics, and harmonization efforts are ongoing within each country and between countries to improve regulatory pathways14–16. These regulatory processes have resulted in the approval of companion diagnostics such as EGFR biomarkers that pair with the EGFR inhibitor afatinib for lung cancer17. Now, international guidelines exist for the use of EGFR and other non-small cell lung cancer companion diagnostics18. The pathways for FDA approval of other biomarkers (that is, biomarkers of risk, diagnosis, response and prognosis not related to a companion therapeutic), designated as Laboratory Developed Tests, have fallen under the FDA purview only recently, and a regulatory framework for these biomarkers is still being developed19. These other types of biomarker have not yet been specifically addressed by the European Union or Japan. With respect to technical validity and reproducibility, the United States requires complex biomarkers to be tested in a Clinical Laboratory Improvement Amendments of 1988 (CLIA)-certified laboratory20, whereas the European Union and Japan do not currently require the use of standardized laboratories for biomarker measurement. The development of regulatory guidelines in even well-developed countries to ensure the safety and utility of biomarkers is an immense effort; thus, the regulation and use of complex biomarkers in less developed countries are likely to continue to lag behind.
The aim of precision medicine and its regulation is to move molecular measurements through validation and ultimately to patient populations in need of improved diagnostic precision. Although the cornerstone of precision medicine will be the large national cohort studies, currently, the precision medicine research approach is being used on existing cohorts and in clinical trials with a much smaller number of individuals. In these studies, large amounts of data from a variety of sources (for example, histology, DNA, protein and RNA) are analysed to determine which pieces of data are the best predictors of disease risk, treatment response and/or prognosis, and thus should be moved forward for validation as biomarkers in future studies. Oncology is a field ripe for using diagnostic tests to subdivide patient populations into risk and treatment categories (that is, new disease subtypes or taxa)2. Lung cancer, in particular, causes more deaths worldwide than the other top three cancers combined21 and therefore presents with a great need for improved diagnostic precision. This Review discusses the biological and statistical strengths and weaknesses of using different types of molecule as biomarkers. Specific examples will be taken from lung cancer to illustrate the potential and pitfalls of each. We then discuss the grouping of different molecule types together to form biomarker panels. Finally, we propose future pathways for precision medicine biomarkers and discuss the potential of biomarkers to stratify patients with lung cancer into different treatment groups to enable precision medicine.
Single-molecule types of biomarker
The measurement of genomics, transcriptomics, pro-teomics, metabolomics, microbiomics and other omics’ methods has now reached a point of reasonable return on investment. The giant reservoirs of molecular data linked to continuously updated electronic medical records envisioned by the Institute of Medicine are now within reach in the foreseeable future. The general precision medicine approach to analysing data is conceptually no different from what has been performed in the past to identify biomarkers in cohort studies. However, it is the proposed magnitude of the amount of data that will need to be collected, stored and analysed to identify the most precise biomarkers possible that is challenging this field (BOX 2).
Box 2 |. The challenges of developing a national precision medicine cohort.
More data collected on more individuals are required to generate validated biomarkers that can improve the precision to which we can categorize an individual’s risk of disease or response to therapy. However, the collection of large amounts of personal data presents complex challenges to researchers, the medical community and individuals whose data are being collected. The challenges facing countries that are developing national cohorts include the following:
Collecting, handling, storing and transporting millions of biospecimens and then analysing these data using multiple different molecular measurement techniques
Collecting electronic medical record data, merging data from different types of medical records and questionnaires, and then storing large amounts of these data
Analysing data from different sources (for example, questionnaires, molecular measurements and electronic medical records) while respecting the strengths and limitations of each type of data
Combining expertise from multiple different disciplines, including clinicians, laboratory researchers, bioinformaticians, biostatisticians and lawyers
Dissemination of these data for researchers to use while ensuring that legal, ethical and privacy concerns of all participants are addressed
The feasibility of national cohorts for the purpose of precision medicine is restricted to countries and regions with the resources to meet all of the above challenges. This requirement will limit the ability of precision medicine to rapidly move to under-resourced regions of the world and its applicability to different races and ethnicities.
This section of the Review focuses on studies that have applied a simplified version of the precision medicine approach, wherein only a single type of molecule (for example, metabolite or protein levels, but not both) in a single biospecimen type is measured in a single cohort. Once identified, putative biomarkers should be validated in other cohorts and a mechanism of action should be elucidated. Once these steps have been accomplished, if the biomarkers were originally measured in tissues that can only be obtained through invasive methods (for example, solid tumour tissue), then blood, urine, saliva and other routes of less invasive specimen extraction should be tested and validated8. Once a feasible platform for measurement is identified (for example, genomic biomarkers may be discovered using next-generation sequencing but often PCR is the preferred platform in the clinic), these biomarkers can then serve to better classify patients in the clinic.
Genomic biomarkers in precision medicine.
Studies of genomic biomarkers in tumour tissue have advanced our understanding of lung adenocarcinoma disease sub-types and taxonomy, particularly at the diagnosis stage. Historically, lung cancer has been grouped into small cell carcinoma, non-small cell squamous cell carcinoma, non-small cell adenocarcinoma and large cell carcinoma subtypes. In the late 1980s and the mid-2000s the research community began to recognize that lung adenocarcinoma could further be subdivided beyond histology into cancers that were driven by KRAS22 and/or EGFR23 gene mutations (FIG. 3). Finally, the development of next-generation DNA sequencing technologies facilitated the comprehensive characterization of the lung cancer genome. Specifically, The Cancer Genome Atlas (TCGA) network has, to date, sequenced approximately 1,025 lung cancer exomes of various histologies and identified at least 15 unique candidate genes that can drive oncogenesis in lung adenocarcinoma when a somatic mutation occurs7,24. New mutations continue to be identified in lung adenocarcinomas such as the recent identification of mutations in protein phosphatase 3 catalytic subunit-α (PPP3CA), histone H3K79 methyl-transferase DOT1L and FtsJ methyltransferase domain containing 1 (PTSJD1; also known as CMTR2)24. Owing to a lack of companion therapeutics and mechanistic studies linkizng newly identified subtypes to disease outcomes, only EGFR-driven or anaplastic lymphoma kinase (ALK)-translocation-driven tumours are treated as different disease subtypes in current clinical practice25,26. Multiple reviews have described the ongoing and promising efforts in the development of genetic biomarkers that are also targets for therapy in lung cancer27–29.
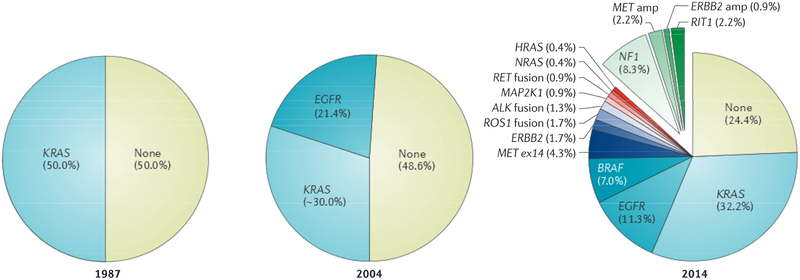
Figure 3|. Knowledge of non-small cell lung adenocarcinoma has evolved in recent decades.
Traditionally, lung cancer was grouped by histology into small cell lung cancer and non-small cell squamous cell carcinoma or adenocarcinoma. In 1987, a KRAS mutation was identified in ~25% of all non-small cell lung cancers, and 50% of lung adenocarcinomas22. In 2004, epidermal growth factor receptor (EGFR) mutations were identified as an additional mutation in lung adenocarcinomas23. The Cancer Genome Atlas (TCGA) Network’s next-generation sequencing of lung adenocarcinoma in 2014 led to the identification of more than 15 different gene events that could be exploited for treatment and/or used for subclassifying patients into new taxa7. ALK, anaplastic lymphoma kinase; amp, amplification; ex, exon; RIT1, Ras like without CAAX1. Data in the left panel were abstracted from Rodenhuis et al.22. Data in the middle panel were abstracted from Paez et al23 and Riley et al.132. The right panel of the figure is from REF. 7, Nature Publishing Group.
The measurement of DNA mutations and translocations as biomarkers has paved the way for further subdivision of lung adenocarcinomas into subtypes that are associated with different outcomes and with different responses to treatment30,31. The best example of these biomarkers currently used in the clinic for patients with lung cancer is the PCR-based companion diagnostic test for EGFR mutations in tumour tissue. This companion diagnostic is used to determine whether the tumour is of the EGFR-positive subtype31; if the patient is EGFR positive, then they can be prescribed an EMA-or FDA-approved EGFR-inhibiting drug (for example, afatinib17,32). Similarly, ALK translocations identified by fluorescent in situ hybridization (FISH) leads to patients being eligible for an EMA-or FDA-approved ALK-inhibiting drug30. Despite sequencing efforts on squamous cell lung cancers33, the identification of new biomarkers and targeted therapy for this type of lung cancer remains limited. However, immunotherapy that targets programmed cell death protein 1 (PD1; also known as PDCD1) is an example of a rare success for this histological subtype34. Many studies have provided evidence that alterations in the non-coding regions of DNA and gene polymorphisms are also associated with disease risk, response or prognosis35–41. The targeting of non-coding regions in particular may be a new avenue of exploration for the prevention and treatment of lung cancer.
Tissue-level DNA measurements are not without their limitations. First, collection of tissue is usually invasive and changes in the DNA are not always functional. Patients with cancer routinely undergo biopsy and tumour removal, which makes this approach feasible despite its invasiveness for initial treatment decisions. However, if subsequent biopsies are needed to make future treatment decisions, then this approach is more challenging. Second, biopsies may not be representative of the whole tumour due to the heterogeneity of multiple malignant cellular clones within a tumour42. Thus, there is an impetus to find less invasive markers of tumour DNA status, for example, measurement of circulating cell-free DNA in lung cancer to identify an increase in EGFR mutations43. However, the low sensitivity of these blood-based measurements will probably limit their use to monitoring, as opposed to diagnosing, lung cancer44. Third, treatment may select for certain cancer clones, as evidenced by the rise in the ratio of EGFR mutations observed in patients with lung adenocarcinoma who had developed resistance to tyrosine kinase inhibitors45. The TRACERx (TRAcking non-small cell lung Cancer Evolution through therapy (Rx)) study46 aims to monitor the impact of tumour heterogeneity on therapeutic outcomes and will address many questions about the response of tumours to treatment over time. The findings from this study are particularly relevant to precision medicine because the behaviour of tumours over time may explain changes in patient outcome, which will be continuously collected from electronic medical records. The invasiveness of biopsies, challenges in identifying functional versus non-functional changes, temporal variation in biomarker values and tumour heterogeneity are not unique to genomic biomarkers. Rather, these factors are challenges to all biomarker development regardless of molecular type.
Transcriptomic biomarkers in precision medicine.
The global measurement of mRNA expression, termed transcriptomics, has provided an understanding of cancer subtypes but, in contrast to DNA, is tissue specific. Evaluation of the transcriptome of non-small cell lung cancer to identify mRNA expression biomarkers began with microarray technology47–50. Using microarray technologies, many studies have identified panels of mRNA expression biomarkers that classify lung cancer into more precise subtypes based on associations with disease outcomes51–55. In contrast to microarrays, which are limited to preselected mRNA probes, mRNA sequencing enables the sequencing of all mRNA present in a sample then, similar to DNA sequencing, maps sequences back to a reference library56. Putative mRNA biomarkers in non-small cell lung cancer are being identified using this newer, more comprehensive approach to searching for biomarkers (for example, Janus kinase (JAK)-signal transduction and activator of transcription (STAT) pathway mRNA57 and mRNA of tumour-educated platelets58) but it is still in preliminary stages.
Epigenomic biomarkers in precision medicine.
DNA methylation, histone protein modifications (for example, methylation and acetylation), microRNA (miRNA) and long non-coding RNA (IncRNA) are all measureable epigenomic biomarkers that function principally to regulate RNAs59,60. miRNA and IncRNA can also have other functions, such as serving as ligands for receptors61, and are measured using the same general methods as mRNA. DNA methylation and histone modifications usually require immunoprécipitation of the epigenomic mark of interest62 or, for DNA methylation, bisulfite conversion63 or restriction enzyme use64 before microarray or sequencing analyses. Epigenomics has been used largely to explore the aetiology of lung cancer, and epigenomic biomarkers are candidate mechanistic biomarkers for classifying individuals based on disease risk. However, as Lilogou et al.59 have recently reviewed, epigenetic biomarkers also have the potential to serve as biomarkers for identifying subclasses of patients with lung cancer. For example, the promoter methylation status of five genes was recently identified as a classifier of non-small cell lung cancer prognosis65. Global methylation patterns, such as CpG island methylator phenotype, have also been associated with prognosis in adenocarcinoma66. Interest in this area continues to grow and there are ongoing clinical trials to move these biomarkers into the clinic.
Proteomic biomarkers in precision medicine.
Immuno-histochemical staining of proteins in formalin-fixed, paraffin-embedded lung tissue samples has been recommended by international experts for use in the clinic to classify tumours67. Studies that used tissue microarrays and existing immunohistochemical protein stains in a high-throughput manner have identified new, putative lung tissue biomarkers68. Advances in mass spectrophotometry that, analogous to next-generation sequencing, enable mapping of a multitude of mass spectrophoto-metric peaks to reference libraries to identify proteins has facilitated the global assessment of the non-small cell lung cancer proteome69,70. Using this new technology, 17 different circulating proteins were recently identified and validated as putative biomarkers for non-small cell lung cancer71. Circulating proteins have also been explored as less invasive biomarkers in lung cancer. Specifically, due to their role in carcinogenesis72, circulating inflammatory proteins have demonstrated clinical utility in lung cancer prognosis73–79. However, moving proteomic biomarkers from the exploratory mass spectrophotometry-based analyses phase into the clinic, which would require more stable measurement platforms, remains challenging. Füzéry et al.80 have thoroughly reviewed such challenges in the framework of the existing FDA approval pathways.
Antigenic proteins expressed on lung cancer and immune cell surfaces are attractive targets for the development of immunotherapeutic antibodies, and have been reviewed extensively81,82. Briefly, intravenous administration of PD1 and PD1 ligand 1 (PDL1) IgG antibodies demonstrate efficacy in the treatment of non-small cell lung cancer. Indeed, two anti-PDl therapies — pembrolizumab83,84 and nivolumab85,86 — have been approved by the EMA and FDA for use in nonsmall cell lung cancer. Recently, PDL1 measurement via immunohistochemistry was approved as a companion diagnostic for pembrolizumab by the FDA87, but the path to approval for this biomarker was fraught with specificity and immunohistochemical challenges88,89. Other attractive immune biomarkers include CD8+ lymphocytes identified by quantitative fluorescence in tumours, which have been associated with better prognosis90. However, tissue imaging-based biomarker identification is hampered by limited throughput and, even with tumour microarrays, often requires a large sample input for a small amount of data output compared with proteomics technology. Nonetheless, the mechanistic relationship between other antigen biomarkers, as companion diagnostics, and antigen-targeting therapies continues to incentivize more research in this rapidly advancing area.
Metabolomic biomarkers in precision medicine.
Similar to proteomics, metabolomics (also known as metabo-nomics) can be assessed in a targeted or unbiased manner, and mass spectrophotometry is used to identify chromatogram peaks as specific metabolites91,92. Metabolomics is particularly promising for biomarker development because altered metabolism is considered a hallmark of cancer72. Moreover, metabolites are frequently exported to the blood for transport or removal from the body via urine or faeces; therefore, these metabolites could serve as non-invasive biomarkers that accurately reflect the metabolic activity of tumour tissues. Tissue93,94, blood95–98 and urinary99,100 metabolomics analyses have yielded putative biomarkers that classify patients into subtypes of lung cancer; however, these findings have yet to be sufficiently validated in more than one cohort while also using positive controls to ensure accurate identification of the purported metabolite. Validation of metabolomic biomarkers requires not only analyses in other cohorts but also a known standard to confirm the identity of the putative metabolite peak. Improved libraries of synthesized standards to authenticate peak identity are an area of need to move this research forward and to build more reliable platforms for metabolite analyses that can progress to the dinic101.
Microbiomic biomarkers in precision medicine.
Using modified extraction procedures, microbial DNA is generally measured in the same manner as human-derived DNA, with the popular exception of 16S rRNA-specific gene sequencing to identify bacteria predominantly at the genus level102,103. The normal lung was thought to be generally sterile until the advancement of culture-independent microbial DNA sequencing techniques for microorganism identification in the lung104,105. As a result of such techniques, we know that cigarette tobacco contains bacteria106 and that cigarette smoke can disrupt the respiratory tract mucosal barrier107 to allow microbiota migration into the lung. These findings have led to the hypothesis that the lung microbiome may play a part in carcinogenesis. To our knowledge, the global analysis of the lung cancer tissue microbiome remains in progress. However, one study has suggested that bacillus species in lung sputum could serve as a non-invasive biomarker of increased lung cancer risk108. The recent identification of Fusobacterium nucleatum as a functional microbial biomarker in colon cancer109–112 provides strong support for the continued interrogation of the microbiome as a functional biomarker with which patients with lung cancer could be classified into risk, response or prognostic subtypes. Antibiotic, probiotic or prebiotic treatment could then be prescribed for different diagnostic sub-types to modify their risk and/or response to therapy113. Standards for faecal microbiome research approaches are emerging114,115, but standards for other biosample types are needed to accelerate the development of non-faecal microbiome biomarkers.
The exposome in precision medicine.
The term ‘exposome’ was first coined by Christopher Wild in 2005 (REF. 116) and refers to all types of molecules and events from the environment to which humans can be exposed; for example, drugs, diet or the microbiome (FIG. 4], Aspects of the exposome are commonly measured by questionnaires, which are administered to patients in the clinic. For lung cancer in particular, information about cigarette smoke and asbestos exposure is requested in addition to information on age and sex to generate a panel of information that is used to stratify people into subtypes of risk of lung cancer117–119 (also see Further information). However, questionnaire responses are biased and error prone. For example, self-reported tobacco exposure does not always correlate with measured tobacco carcinogen exposures120. Thus, development of molecular biomarkers of tobacco smoke121,122 and other exposures that could be used in the clinic to more accurately reflect a patient’s expo-some, and therefore lung cancer risk classification, are ongoing.
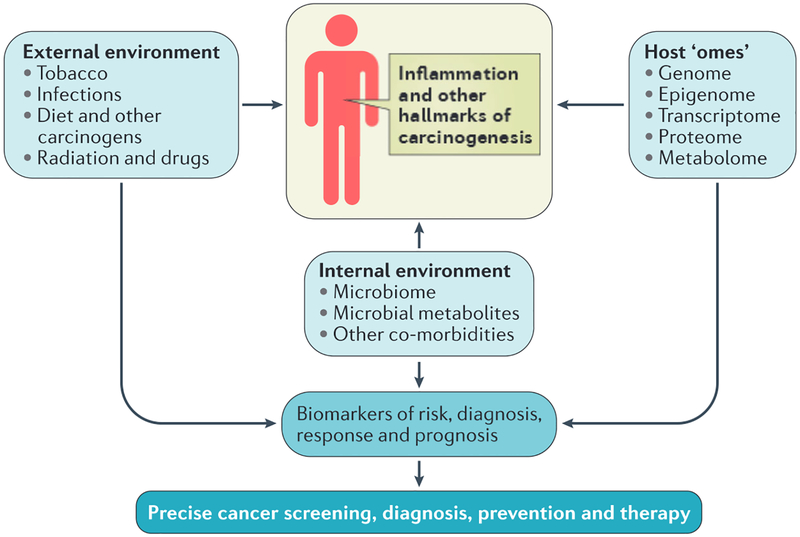
Figure 4|. The lung exposome.
The exposome of the lung comprises a diverse array of molecules and events (including carcinogens from tobacco, asbestos and radon) that come from the external and internal lung environment. These external and internal influences interact with each other and host’omes’to alter the lung cell environment (including inflammation and the microbiome) and may promote or protect against the development of the hallmarks of cancer72. Smoking is estimated to cause 90% of lung cancers. Occupational exposures to carcinogens and radon exposure are estimated to cause 9–15% and 10% of lung cancer cases, respectively174. Measurement of the exposome, in addition to other host ‘omics’, has led to the development of precise biomarkers of risk, diagnosis, treatment response and prognosis by which patients can be classified into new taxa. These new taxa then require different standards of care for cancer screening, diagnosis, prevention and therapy.
Interestingly, the exposome alters the effects of other molecular measurements (for example, inflammation) and the effects of the exposome may be altered by changes in other molecules such that it is imperative that the exposome be considered when developing any type of biomarker. Specifically, among patients with lung adenocarcinoma, smokers have a higher mutation frequency overall than non-smokers123, and it is known that exposure to smoke can cause gain-of-function TP53 mutations124,125, which drive lung carcinogenesis. Although questionnaire-derived data have been analysed predominantly by epidemiologists and molecular data by basic scientists, these data are more commonly being incorporated together in research projects, and collaboration across disciplines is essential to ensure these data are analysed to their fullest potential. The combination of the exposome and molecular biomarkers for improved prediction of disease subclassifications is discussed in more detail below.
Summary.
Nearly all omics’ analyses are still limited by factors that have slowed the progression of biomarkers from discovery to deployment in the clinic. The following are the most important factors impeding progress. First, there are technical reproducibility issues of ‘omics’ platforms and variability between laboratories126, because the vast majority of biomarker studies are not conducted in CLIA-certified or other regulated laboratories. Second, there are limitations in the quality and size of the reference library used to identify molecules. Third, false positives due to the vast amount of potential biomarkers analysed in global omics’ studies127,128. Fourth, statistical reproducibility issues arise owing to biases in the original sample or validation cohort, or owing to false discoveries8,129. Fifth, lack of longitudinal cohorts in which to validate biomarkers over time. Sixth, the need for functional studies of putative biomarkers. Finally, heterogeneity within and between samples leads to inconsistent measurements on the same sample or measurements that are out of the dynamic range of a test, respectively.
These limitations have led to a plethora of putatively identified biomarkers in the literature that lack validation, mechanistic evidence and/or follow-up studies. Moreover, many biomarkers are identified at the tissue level. Although tissue-level studies are often crucial in identifying a mechanistic link between a biomarker and carcinogenesis, biomarkers requiring biopsy are not practical for assessing cancer risk and for monitoring response to treatment in the dinic, even in well-developed health-care systems. Perhaps the expansion of regulations regarding laboratory developed tests in the United States19, and potentially the European Union and Japan, will incentivize the biomedical community to move putatively identified biomarkers towards less invasive, validated biomarkers not only for companion diagnostics but also for biomarkers of disease prognosis and risk.
The ability of any biomarker to correctly differentiate two subgroups of patients with lung cancer in a statistically significant and clinically meaningful manner is dependent on the relative number of people in each group and the magnitude of the difference in the value and variance of the biomarker in each group (that is, power)127–129. Furthermore, all analyses comparing two lung cancer subgroups assumes that the groups are identical to each other in all other ways except for the biomarker or biomarkers that differentiate the lung cancer subtype130. Thus, the rarer a disease subtype is the less likely it will be that even large cohorts will have the power (number of participants and measured differences between subgroups) to meet the assumptions to identify these subgroups. Nonetheless, innovative clinical trial design approaches (for example, n = 1 studies131) are leading the way for the approval of companion diagnostics and are helping to minimize the challenge of not observing enough participants in each subgroup27. In addition, the more studies that are conducted and the more sensitive or specific molecular measurement platforms and procedures become the more statistical power we will have to identify and validate new biomarkers. These new approaches, paired with the precision medicine approach6 to continuously re-evaluate the outcomes for patients classified to a disease subtype by biomarkers in a longitudinal manner, will change the way biomarkers are developed and will enable the most precise classification of patients in the future.
Multi-molecule-types of biomarker panel
The Institute of Medicine’s seminal report envisioned an Information Commons that contains multiple omics’ approaches such that biomarker panels containing different molecule types, exposome data and/or demographic data could be developed to classify diseases into more precise subtypes6. Although single-molecule biomarkers (for example, EGFR) have progressed into the clinic31, biomarker panels are still in the discovery stage. Biomarker panels containing various types of molecule are attractive because genes, proteins, RNAs and metabolites all work in concert to prevent or promote the development of the hallmarks of cancer72. However, the development of biomarker panels is still limited by two main factors. First, by any weaknesses associated with each individual omics’ technique, molecule type and tissue type included in the panel, and second, by amplification of the factors identified as challenges common to the identification of any biomarker mentioned above8,127–129. Integrating different types of data leads to more potential biomarker combinations and, consequently, more functional studies and statistical tests that need to be run. These additional statistical tests require more power to detect significant differences while still trying to avoid false positives127–129. Two conceptual approaches to developing biomarker panels have been used. The first relies on adding new biomarkers to existing biomarkers or biomarker panels to improve the sensitivity or specificity of the panel because of either an interaction or an independent effect of the new biomarker. The second approach employs de novo analysis and integration of multiple sources of molecular data to identify the best combination of putative biomarkers.
Adding new biomarkers to existing biomarker panels in precision medicine.
The addition of new biomarkers to existing, validated biomarker, demographic and exposome information (for example, smoking status, age, race and sex) panels decreases the number of possible biomarker combinations. Thus, the statistical power needed to identify additional biomarkers compared with de novo analysis (discussed below) is also decreased as compared with de novo analyses. An early, seminal example of adding a biomarker to existing predictive information in lung cancer was the discovery that smoking (exposome) can cause TP53 mutations and thus alters lung cancer risk124. However, treatments targeting p53 or KRAS pathways (KRAS is also frequently mutated7 and associated with smoking in lung cancer132) remain in clinical trial stages. Interestingly, most of the work in this area was conducted before advanced next-genera-tion sequencing and used exhaustive mechanistic studies to identify this relationship. Conversely, the majority of newer biomarker studies pair epidemiological evidence with global ‘omics’-based technology, and then, if validated, few have identified a mechanism of action that explains the relationship between these biomarkers and disease. Identifying a mechanism of action is required for the development of viable companion therapeutics.
The precision medicine research approach has been used in lung cancer to interrogate whether the addition of new molecular data improves an existing biomarker panel. A recent publication using a TCGA dataset has provided evidence that the addition of single-molecule type biomarkers (copy number alterations, protein and miRNA measurements) to existing exposome-only predictors improves prognostic accuracy in lung squamous cell carcinoma133. With respect to early-stage lung adenocarcinoma, mir-21 methylation134, homeobox A9 (HOXA9) methylation135 and the expression levels of a panel of four genes55 have been validated in multiple cohorts to independently predict lung cancer outcomes. However, the addition of mir-21 (REF. 51) and HOXA9 methylation status135 to the gene expression biomarker panel improves the predictive accuracy above any of these biomarkers alone55. Analyses of multiple international cohorts have provided evidence that the risk of lung cancer from asbestos exposure is increased by the presence of certain genetic variants136. Although not multi-omic’, there are ongoing clinical trials that use traditional histological subtyping and then use genomic analysis in patients with lung cancer using a variety of platforms. Examples of such trials include the BATTLE-2 trial137 and a US National Cancer Institute trial138. These results are then used to make treatment decisions and to identify de novo genetic variations associated with disease response and/or prognosis as candidate biomarkers. These studies are the pinnacle of precision medicine research as it currently exists and, if successful, will change clinical practice within the next decade. This relatively straightforward approach of adding new biomarkers requires a thoroughly validated existing biomarker or panel of biomarkers and could possibly miss unique combinations of biomarkers that may serve as better predictors. With the recent improvements in computational technologies, there has also been a movement towards de novo analysis of multiple omics’ datasets to truly integrate the data and identify novel biomarker panels.
De novo analysis combining global datasets to generate biomarker panels in precision medicine.
Integrating multiple datasets, often derived from global omics’ analyses of different types of molecule, for biomarker panel development has largely been stymied by a lack of methodological approaches that are suitable for combining different data sources. Recent advances in network analysis and other mathematical modelling approaches are swiftly moving this field forward139–143; however, no preferred approach has emerged. Using TCGA data, Li et al,144 integrated genomic, transcriptomic and proteomic information to classify patients with non-small cell lung cancer by prognosis. Kim et al.145 integrated DNA, mRNA, miRNA and methylation sequencing data to identify putative biomarkers that classify female patients with non-smoking-associated lung adenocarcinoma into distinct subtypes. However, identifying mechanisms of action and finding cohorts with sufficient sample size (with meaningful racial, ethnic and geographic diversity) in which to validate these, and other, de novo-assembled biomarker panels remain major challenges. The Precision Medicine Initiative cohort1,2, the UK Biobank4, The International Cancer Genome Consortium146 and other large studies with molecular measurement data will provide an unparalleled opportunity for validating biomarker panels owing to the projected collection of multiple ‘omics’ data that is anticipated to be publicly available for researchers.
Summary.
Putative biomarker panels in lung cancer are just beginning to accrue55,133,144,145, but their path to the clinic will probably be even longer than for singlemolecule biomarkers owing to exacerbation of typical challenges associated with biomarker development by combining ‘omics’ approaches. Specifically, combining biomarkers leads to multiple potential combinations that require the statistical power to be tested. Using different types of molecule in one panel also leads to logistical challenges on how to measure different molecule types (for example, proteins and DNA) on the same platform when moving towards regulatory approval for clinical use. Nonetheless, biomarker panels are hypothesized to provide a more realistic picture of aberrant regulation in complex diseases such as cancer because molecules do not function in isolation to generate a phenotype. Small shifts in the relative amounts of RNA, protein, epigenetic modifications, metabolites and microorganisms over time may also be useful for early prediction of disease risk or outcomes. For example, a budding area of research in precision medicine is network medicine. Network medicine involves the use of emerging network approaches to integrate omics’ measurements139–143 to look for small changes over time in the relationship between ‘omics’ that are associated with disease143,147–149. Such dynamic analysis is in contrast to the current use of static panels of biomarkers. This continual monitoring approach is consistent with the greater vision of precision medicine; however, it should rely on only minimally invasive biomarkers and therefore lends itself well to the use of microfluidics150.
Summary and conclusions
The reduced cost and increased reproducibility of new ‘omics’ technologies, new methodological approaches for integrating different types of molecular data139–143, and the number of publicly available datasets with molecular measurements have all been pivotal in achieving the substantial leaps forward in biomarker development that we have witnessed over the past decade (BOX 3). The precision medicine research approach is simply a faster paced, larger scale, integrated version of the traditional, single measurement-based biomarker development approach that has only been made possible due to these advances. Currently, there are large prospective, international cohort studies (for example, the European Prospective Investigation into Cancer and nutrition (EPIC)151 and the Women’s Health Initiative152) and electronic medical record-based datasets (for example, electronic MEdical Records and GEnomics (eMERGE)153 and the UK Biobank4) with biobanked samples that allow for ongoing biomarker discovery. The number of biomarkers moving from discovery to clinical trials is worryingly small. Furthermore, the vast majority of registered clinical trials test biomarkers as companion diagnostics to treatment because there are existing regulatory frameworks for companion diagnostics whereas biomarker panels for risk of disease and prognosis (which are unrelated to a specific treatment) have murkier regulatory pathways.
Box 3 |. Promising biomarkers in lung cancer.
A summary of the promising biomarkers in lung cancer is provided below. For more detail, please refer to Lung Cancer and Personalized Medicine: Novel Therapies and Clinical Management as part of the Advances in Medicine and Biology series (2016)173.
Tumour immune and microenvironment biomarkers. For example, programmed cell death protein 1 (PD1), PD1 ligand 1 (PDL1) and vascular endothelial growth factor A (VEGFA)
Genetic aberration biomarkers. For example, KRAS, HER2 (also known as ERBB2), BRAF, MET, ROSI, RET, fibroblast growth factor receptor 1 (FGFR1), SRY-BOX 2 (SOX2), platelet-derived growth factor receptor-a (PDGFRA), discoidin domain receptor tyrosine kinase 2 (DDR2), PI3K catalytic subunit-a(P/K3CA), PTEN, mixed lineage leukaemia 2 (MLL2; also known as KMT2D)
Epithelial-to-mesenchymal transition-associated biomarkers. For example, SLUG, forkhead box C2 (FOXC2) and transforming growth factor-β (TGFβ)
Resistance and susceptibility to treatment biomarkers. For example, ERCCl, ribonucleoside-diphosphate reductase (RRM) and thymidylate synthase (TS)
In addition to changes in regulations, the field of biomarker discovery has innate practical challenges. Specifically, the precision of disease subtyping by biomarkers to predict risk, response or prognosis is limited by a high risk of false positives when seeking to identify a biomarker from the global measurement of thousands of molecules. Moreover, questions of basic statistical power (that is, the number of patients presenting with a disease subtype)8,127,129, the need for validation and functional studies, and the follow-through to develop non-invasive biomarkers are also limitations. McShane et al.154–162 have published a series of manuscripts on REporting recommendations for tumour MARKer prognostic studies (REMARK). Consistent adherence to these publishing guidelines will enable improved transparency and allow the scientific community to better judge the quality of the plethora of studies reporting putative biomarker identification. Importantly, the success of these complex studies will require effective collaborative science; a good example is the success demonstrated by the Early Detection Research Network at the US National Cancer Institute163. The rate at which new biomarkers enter the clinic will now be benchmarked by the requirements set forth by regulatory agencies9–16,19. Changes in regulation that result in profitability for biomarkers not tied to a specific drug (for example, biomarker panels to estimate disease risk), will serve to spur the movement of biomarkers from lone studies to the clinic. The speed of discovery and validation of biomarkers could be improved by real-time data collection, which would, theoretically, allow for faster monitoring and revision of new disease taxa as data on patients are collected. However, this benefit must be balanced against the cost of screening a large population multiple times and the increased risk of false positives (over-diagnosis) simply due to the number of measurements being undertaken.
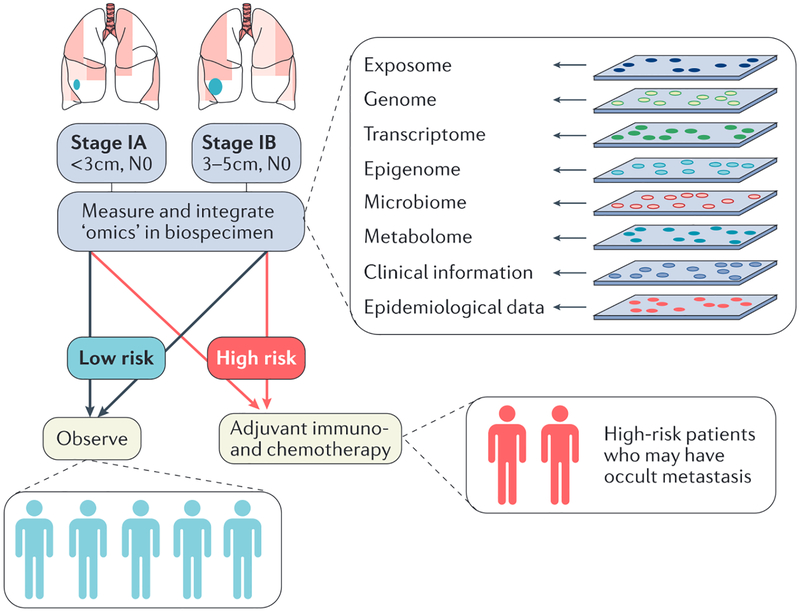
Analyses of existing cohorts and the generation of national cohorts (that is, in the United States1,2, the United Kingdom4, and in Denmark5 and Germany5) promises to address many of the statistical and logistical concerns in biomarker development, allowing for the progress of truly precise medicine. One can envision that machine learning could be used to mine and continually improve algorithms for calculating patient risk, diagnosis, response and prognosis over time. These algorithms could then be applied to the general population to ensure patients receive appropriate care while measuring only the necessary biomarkers at key time points identified from national cohorts. Furthermore, with respect to the heterogeneous nature of cancers, there may be a day in the future when a tumour is profiled and then a follow-up biomarker panel and drug are developed in real time specifically for a single tumour. Although these possibilities are only likely to be realized decades in the future, currently, the precision medicine approach has led to the identification of new subtypes of non-small cell lung cancers (EGFR, ALK, TP53 and KRAS) and the translation of companion diagnostic biomarkers (for EGFR and ALK) to diagnose and choose appropriate treatment regimens for these new subtypes in the clinic31. However, there is an ongoing need for translatable biomarkers of disease risk, particularly for smokers, and prognosis for patients with EGFR-negative and ALK-negative lung adenocarcinomas. There is currently a large screening trial being conducted in the United Kingdom to test the efficacy of low-dose computed tomography (LDCT) before initiating a national lung cancer screening programme164. LDCT has recently been approved for use in screening individuals at high risk for lung cancer in the United States165,166, and its implementation will lead to an increased number of individuals diagnosed with early-stage lung cancer. An estimated 8.6 million to 8.8 million people in the United States meet the current criteria for LDCT167,168 and, if they are screened, this will lead to an overwhelming number of true and false positive findings that will require treatment decisions165,166,169. This large number is because, despite ongoing efforts to improve the specificity of imaging-based biomarkers170, the false-positive rate for LDCT is estimated at >90% (REF. 165). Thus, one especially high impact area for the development of precision medicine biomarkers is to further classify early-stage (IA and IB) lung cancers into subtypes of patients at high risk for cancer recurrence to inform treatment decisions171 (FIG. 5). A similar approach would be useful in differentiating findings from mammography, which are also plagued by high false-positive rates172. This improved specificity would then allow oncologists to treat the patients who would benefit and minimize overtreatment of those who are unlikely to progress. This example is a clear demonstration of the power of precision medicine biomarkers and the future of medicine.
Figure 5 |. Use of precision medicine to classify patients with early-stage lung cancer into subclasses to provide appropriate treatment.
Approximately 25% of patients with stage I lung cancer will have recurrent disease associated with occult metastasis. This figure depicts the classification of early stage (IA and IB) lung cancers by a single biomarker or a panel of biomarkers that predicts risk of recurrence generated using a precision medicine research strategy into’low risk for recurrence’and ‘high risk for recurrence’. Once classified into subclasses (taxa), low-risk patients can be observed post-curative surgery whereas high-risk patients can be provided options for adjuvant therapy post-surgery.
Acknowledgements
This work was supported by funding from the Intramural Program of the Center for Cancer Research, National Cancer Institute, Bethesda, Maryland, USA, and the Cancer Prevention Fellowship Program, National Cancer Institute, Rockville, Maryland, USA.
Footnotes
Competing interests statement
The authors declare no competing interests.
DATABASES
Cancer of the Lung Evaluation and Assessment of Risk (CLEAR): http://www3.mdandenderson.org/depts/epidemiology/spitzLCtool/controller.cfc?method=getSubjectData Lung Cancer Screening DecisionTool: http://nomogrgms.mskcc.org/Lung/Scregning.aspx
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Obama B Remarks by the President in State of the Union Address | January 20, 2015. The White House https://www.whitehouse.gov/the-press-office/2015/01/20/remarks-president-state-union-address-january-20-2015 (2015). [Google Scholar]
- 2.Collins FS & Varmus H A new initiative on precision medicine. N. Engl. J. Med 372, 793–795 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; An overview of the US approach to generate a national cohort for precision medicine research.
- 3.US National Institutes of Health (NIH). NIH workshop on building a precision medicine research cohort NIH http://www.nih.gov/precisionmedicine/wprKshop.htm (2015). [Google Scholar]
- 4.Elliott P, Peakman TC & Biobank UK. The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int. J. Epidemiol 37, 234–244 (2008). [DOI] [PubMed] [Google Scholar]; A summary of the UK Biobank, which will serve as an important resource for precision medicine research.
- 5.Frank L Epidemiology. When an entire country is a cohort. Science 287, 2398–2399 (2000). [DOI] [PubMed] [Google Scholar]
- 6.National Research Council. Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease (National Academies Press, 2011). [PubMed] [Google Scholar]; A comprehensive review of precision medicine research, a taxonomy of terms and a suggested approach for conducting precision medicine research in the future.
- 7.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive analysis of lung adenocarcinoma describing many different molecular subtypes.
- 8.Pepe MS et al. Phases of biomarker development for early detection of cancer. J. Natl Cancer Inst 93, 1054–1061 (2001). [DOI] [PubMed] [Google Scholar]; A summary of common concerns when developing biomarkers for clinical use.
- 9.US Food and Drug Administration (FDA). In Vitro Companion Diagnostic Devices: Guidance for Industry and Food and Drug Administration Staff (CBER/CDRH/FDA, 2014). [Google Scholar]
- 10.Mansfield EA FDA perspective on companion diagnostics: an evolving paradigm. Clin. Cancer Res 20, 1453–1457 (2014). [DOI] [PubMed] [Google Scholar]
- 11.European Medicines Agency (EMEA). Definitions for Genomic Biomarkers, Pharmacogenomics, Pharmacogenetics, Genomic Data and Sample Coding Categories (EMEA/CHMP/ICH, 2007). [Google Scholar]
- 12.European Medicines Agency (EMA). Qualification of Novel Methodologies for Drug Development: Guidance to Applicants (EMA/CHMP/SAWP, 2009). [Google Scholar]
- 13.Pharmaceuticals and Medical Devices Agency. Technical Guidance on Development of In Vitro Companion Diagnostics and Corresponding Therapeutic Products (Ministry of Health, Labour and Welfare, 2013). [Google Scholar]
- 14.Pignatti F et al. Cancer drug development and the evolving regulatory framework for companion diagnostics in the European Union. Clin. Cancer Res 20, 1458–1468 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Senderowicz AM & Pfaff O Similarities and differences in the oncology drug approval process between FDA and European Union with emphasis on in vitro companion diagnostics. Clin. Cancer Res 20, 1445–1452 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Tazawa Y Perspective for the development of companion diagnostics and regulatory landscape to encourage personalized medicine in Japan. Breast Cancer 23, 19–23 (2015). [DOI] [PubMed] [Google Scholar]
- 17.US Food and Drug Administration (FDA). FDA approves afatinib FDA http://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm560574.him (2016). [Google Scholar]
- 18.Lindeman NI et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Mol. Diagn 15,415–453 (2013). [DOI] [PubMed] [Google Scholar]
- 19.McNeil C Regulating laboratory-developed tests: devil will be in details. J. Natl Cancer Inst 107, djvl 13 (2015). [DOI] [PubMed] [Google Scholar]
- 20.US Food and Drug Administration (FDA). Administrative Procedures for CUA Categorization: Guidance for Industry and Food and Drug Administration Staff (CDRH/DHHS/FDA, 2014). [Google Scholar]
- 21.American Cancer Society. Cancer Facts & Figures 2014. (American Cancer Society, 2014). [Google Scholar]
- 22.Rodenhuis S et al. Mutational activation of the K~ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N. Engl. J. Med 317, 929–935(1987). [DOI] [PubMed] [Google Scholar]
- 23.Paez JG et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Campbell JD et al. Distinct patterns of somatic genome alternations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet 48,607–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerr KM et al. Second ESMO consensus conference on lung cancer: pathology and molecular biomarkers for non-small-cell lung cancer. Ann. Oncol 25, 1681–1690 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Korpanty GJ, Graham DM, Vincent MD & Leighl NB Biomarkers that currently affect clinical practice in lung cancer: EGFR, ALK, MET, ROS-1, and KRAS. Front. Oncol 4, 204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimou A & Papadimitrakopoulou V Non-small cell lung cancer beyond biomarkers: the evolving landscape of clinical trial design. J. Pers. Med 4, 386–401 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; A description of new efforts to adapt clinical trials to address precision medicine questions in lung cancer.
- 28.Kumar M, Emani V & Owonikoko TK Biomarkers and targeted systemic therapies in advanced non-small cell lung cancer. Mol. Aspects Med 45, 55–66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rooney C & Sethi T Advances in molecular biology of lung disease: aiming for precision therapy in non-small cell lung cancer. Chest 148, 1063–1072 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Kwak EL et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med 363, 1693–1703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallee A, Le Loupp AG & Denis MG Efficiency of the Therascreen(R) RGQ PCR kit for the detection of EGFR mutations in non-small cell lung carcinomas. Clin. Chim. Acta 429, 8–11 (2014). [DOI] [PubMed] [Google Scholar]
- 32.European Medicines Agency (EMA). Giotrib (afatinib) EMA http://www.ema.europa.eu/ema/index.jsp?curl-pages/medicines/human/medicines/002280/humanmed001698.jsp&mid-WC0b01ac058001d124(2016). [Google Scholar]
- 33.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Topalian SL et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med 366, 2443–2454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butkiewicz D et al. Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis 22, 593–597 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Li Y et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 11, 321–330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mechanic LE et al. Common genetic variation in TP53 is associated with lung cancer risk and prognosis in African Americans and somatic mutations in lung tumors. Cancer Epidemiol. Biomarkers Prev. 16,214–222 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Mechanic LE et al. Polymorphisms in XPD and TP53 and mutation in human lung cancer. Carcinogenesis 26, 597–604 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Pine SR et al. Lung cancer survival and functional polymorphisms in MBL2, an innate-immunity gene. J. Natl Cancer Inst 99, 1401–1409 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan BM et al. Identification of a functional SNP in the 3’UTR of CXCR2 that is associated with reduced risk of lung cancer. Cancer Res. 75, 566–575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang L et al. Association between single nucleotide polymorphisms (SNPs) and toxicity of advanced nonsmall-cell lung cancer patients treated with chemotherapy. PLoS ONE 7, e48350 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerlinger M et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med 366, 883–892 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mazurek A et al. Quantification of concentration and assessment of EGFR mutation in circulating DNA. Cancer Biomark. 15, 515–524 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Luo J, Shen L & Zheng D Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci. Rep 4, 6269 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pao W et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoSMed. 2, e73 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jamal-Hanjani M et al. Tracking genomic cancer evolution for precision medicine: the lung TRACERx study. PLoS Biol. 12, el001906 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beer DG et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med 8, 816–824 (2002). [DOI] [PubMed] [Google Scholar]
- 48.Bhattacharjee A et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl Acad. Sci. USA 98, 13790–13795 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garber ME. et al. Diversity of gene expression in adenocarcinoma of the lung. Proc. Natl Acad. Sci. USA 98, 13784–13789 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kikuchi T et al. Expression profiles of non-small cell lung cancers on cDNA microarrays: identification of genes for prediction of lymph-node metastasis and sensitivity to anti-cancer drugs. Oncogene 22, 2192–2205 (2003). [DOI] [PubMed] [Google Scholar]
- 51.Akagi I et al. Combination of protein coding and noncoding gene expression as a robust prognostic classifier in stage I lung adenocarcinoma. Cancer Res. 73,3821–3832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen HY et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N. Engl. J. Med 356, 11–20 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Hadara H et al. A five-gene and corresponding protein signature for stage-1 lung adenocarcinoma prognosis. Clin. Cancer Res 17, 1490–1501 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee ES et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin. Cancer Res 14, 7397–7404 (2008). [DOI] [PubMed] [Google Scholar]
- 55.Okayama H et al. The expression of four genes as a prognostic classifier for stage I lung adenocarcinoma in 12 independent cohorts. Cancer Epidemiol. Biomarkers Prev. 23, 2884–2894 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oshlack A, Robinson MD & Young MD From RNA-seq reads to differential expression results. Genome Biol. 11, 220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Govindan R et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 150, 1121–1134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Best MG et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 28, 666–676(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liloglou T, Bediaga NG, Brown BR, Field JK & Davies MP Epigenetic biomarkers in lung cancer. Cancer Lett. 342, 200–212 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Schmitt AM & Chang H Y Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fabbri M et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl Acad. Sci. USA 109, E2110–E2116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Massie CE & Mills IG Chromatin immunoprecipitation (ChIP) methodology and readouts. Methods Mol. Biol 505, 123–137 (2009). [DOI] [PubMed] [Google Scholar]
- 63.Herman JG, Graff JR, Myohanen S, Nelkin BD & Baylin SB Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA 93, 9821–9826 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schumacher A, Weinhausl A & Petronis A Application of microarrays for DNA methylation profiling. Methods Moi. Biol 439, 109–129 (2008). [DOI] [PubMed] [Google Scholar]
- 65.Sandoval J et al. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J. Clin. Oncol 31, 4140–4147 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Shinjo K et al. Integrated analysis of genetic and epigenetic alterations reveals CpG island methylator phenotype associated with distinct clinical characters of lung adenocarcinoma. Carcinogenesis 33, 1277–1285 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Travis WD et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol 6, 244–285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brunnstrom H et al. Immunohistochemistry in the differential diagnostics of primary lung cancer: an investigation within the Southern Swedish Lung Cancer study. Am. J. Clin. Pathol 140, 37–46 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Yanagisawa K et al. Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 362, 433–439 (2003). [DOI] [PubMed] [Google Scholar]
- 70.Zhukov TA, Johanson RA, Cantor AB, Clark RA STockman, M. S. Discovery of distinct protein profiles specific for lung tumors and pre-malignant lung lesions by SELD1 mass spectrometry. Lung Cancer 40, 267–279 (2003). [DOI] [PubMed] [Google Scholar]
- 71.Kim YJ et al. Verification of the biomarker candidates for non-small-cell lung cancer using a targeted proteomics approach. J. Proteome Res 14, 1412–1419 (2015). [DOI] [PubMed] [Google Scholar]
- 72.Hanahan D & Weinberg RA Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]; A summary of the hallmarks, or characteristics, of cancer.
- 73.Alifano M et al. Preresection serum C-reactive protein measurement and survival among patients with resectable non-small cell lung cancer. J. Thorac. Cardiovasc. Surg 142, 1161–1167 (2011). [DOI] [PubMed] [Google Scholar]
- 74.Barrera L et al. Cytokine profile determined by data-mining analysis set into clusters of non-small-cell lung cancer patients according to prognosis. Ann. Oncol 26, 428–435 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Enewold L et al. Serum concentrations of cytokines and lung cancer survival in African Americans and Caucasians. Cancer Epidemiol. Biomarkers Prev 18, 215–222 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hong S, Kang YA, Cho BC & Kim DJ Elevated serum C-reactive protein as a prognostic marker in small cell lung cancer. Yonsei Med. J 53, 111–117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ryan BM, Pine SR, Chaturvedi AK, Caporaso N & Harris CC A combined prognostic serum interleukin-8 and interleukin-6 classifier for stage 1 lung cancer in the prostate, lung, colorectal, and ovarian cancer screening trial. J. Thorac. Oncol 9, 1494–1503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Song G et al. Personalized biomarkers to monitor disease progression in advanced non-small-cell lung cancer patients treated with icotinib. Clin. Chim. Acta 440, 44–48 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Srimuninnimit V et al. C-reactive protein as a monitor of chemotherapy response in advanced non-small cell lung cancer (CML study). J. Med. Assoc. Thai 95 (Suppl. 2), S199–S207 (2012). [PubMed] [Google Scholar]
- 80.Füzéry AK, Levin J, Chan MM & Chan DW Translation of proteomic biomarkers into FDA approved cancer diagnostics: issues and challenges. Clin. Proteom 10, 13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Atkins MB & Larkin J Immunotherapy combined or sequenced with targeted therapy in the treatment of solid tumors: current perspectives. J. Natl Cancer Inst 108, djv414 (2016). [DOI] [PubMed] [Google Scholar]; A review of the potential to combine precisely targeted therapies with immunotherapies in oncology.
- 82.Topalian SL, Taube JM, Anders RA & Pardoll DM Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 16, 275–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.European Medicines Agency (EMA). Keytruda (pembrolizumab) EMA http://www.ema.europa.eu/ema/index.jsp?curl-pages/medicines/human/medicines/005820/humanmed001886.jsp&mid=WC0b0Iac058001dl24 (2015) [Google Scholar]
- 84.US Food and Drug Administration (FDA). FDA approves Keytruda for advanced non-small cell lung cancer FDA http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm465444.htm (2015). [Google Scholar]
- 85.US Food and Drug Administration (FDA). FDA expands approved use of Opdivo to treat lung cancer FPA http://www.fda.gov/Nev/sEvents/Newsroom/PressAnnouncements/ucm436534.htm (2015). [Google Scholar]
- 86.European Medicines Agency (EMA). Nivolumab BMS (nivolumab) EMA http://www.ema.europa.eu/ema/index.jsp?curl=pages/msdicines/human/medicines/005840/humanmed001887.jsp&mid=WC0b013C058001d124 (2016). [Google Scholar]
- 87.US Food and Drug Administration (FDA). PHARMDX premarket approval FDA http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfPMA/pma.cfm?id-P150013 (2015). [Google Scholar]
- 88.Patel SP & Kurzrock R PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Ther 14, 847–856 (2015). [DOI] [PubMed] [Google Scholar]
- 89.Shukuya T & Carbone DP Predictive markers for the efficacy of anti-PD-l/PD-Ll antibodies in lung cancer. J. Thorac. Oncol, 10.1016/j.jtho.2016.02.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schalper KA et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J. Natl Cancer Inst 107, dju435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nicholson JK, Connelly J, Lindon JC & Holmes E Metabonomics: a platform for studying drug toxicity and gene function. Nat. Rev. Drug Discov 1, 153–161 (2002). [DOI] [PubMed] [Google Scholar]
- 92.Wishart DS Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov 10.1058/nrd.2016.52 (2016). [DOI] [PubMed] [Google Scholar]
- 93.Rocha CM et al. NMR metabolomics of human lung tumours reveals distinct metabolic signatures for adenocarcinoma and squamous cell carcinoma. Carcinogenesis 36, 68–75 (2015). [DOI] [PubMed] [Google Scholar]
- 94.Wikoff WR et al. Metabolomic markers of altered nucleotide metabolism in early stage adenocarcinoma. Cancer Prev. Res. (Phila.) 8, 410–418(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fahrmann JF et al. Investigation of metabolomic blood biomarkers for detection of adenocarcinoma lung cancer. Cancer Epidemiol. Biomarkers Prev. 24, 1716–1723 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hori S et al. A metabolomic approach to lung cancer. Lung Cancer 74, 284–292 (2011). [DOI] [PubMed] [Google Scholar]
- 97.Li Y, Song X, Zhao X, Zou L & Xu G Serum metabolic profiling study of lung cancer using ultra high performance liquid chromatography/quadrupole time-of-flight mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 966, 147–153 (2014). [DOI] [PubMed] [Google Scholar]
- 98.Miyamoto S et al. Systemic metabolomic changes in blood samples of lung cancer patients identified by gas chromatography time-of-flight mass spectrometry. Metabolites 5, 192–210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Carraia J et al. Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of urine. J. Proteome Res 10, 221–230 (2011). [DOI] [PubMed] [Google Scholar]
- 100.Mathe EA et al. Noninvasive urinary metabolomic profiling identifies diagnostic and prognostic markers in lung cancer. Cancer Res. 74, 3259–3270 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Castle AL, Fiehn O, Kaddurah-Daouk R & Lindon JC Metabolomics Standards Workshop and the development of international standards for reporting metabolomics experimental results. Brief. Bioinformat 7, 159–165 (2006). [DOI] [PubMed] [Google Scholar]
- 102.Handelsman J, Rondon MR, Brady SF, Clardy J & Goodman RM Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol 5, R245–R249 (1998). [DOI] [PubMed] [Google Scholar]
- 103.Kozich JJ, Westcott SL, Baxter NT, Highlander SK & Schloss PD Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appi. Environ. Microbiol 79, 5112–5120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pragman AA, Kim HB, Reilly CS, Wendt C & Isaacson RE The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS ONE 7, e47305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Segal LN & Blaser MJ A brave new world: the lung microbiota in an era of change. Ann. Am. Thorac. Soc 11 (Suppl. 1), S21–S27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sapkota AR, Berger S & Vogel TM Human pathogens abundant in the bacterial metagenome of cigarettes. Environ. Health Perspect 118, 351–356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heijink IH, Brandenburg SM, Postma DS & van Oosterhout AJ Cigarette smoke impairs airway epithelial barrier function and cell-cell contact recovery. Eur. Respir. J 39, 419–428 (2012). [DOI] [PubMed] [Google Scholar]
- 108.Hosgood HD et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ. Mol. Mutag 55, 643–651 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Castellarin M et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Flanagan L et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis 33, 1381–1390 (2014). [DOI] [PubMed] [Google Scholar]
- 111.Kostic AD et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kostic AD et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zitvogel L, Ayyoub M, Routy B & Kroemer G Microbiome and anticancer immunosurveillance.Cell 165, 276–287 (2016). [DOI] [PubMed] [Google Scholar]
- 114.Cardona S et al. Storage conditions of intestinal microbiota matter in metagenomic analysis. BMC Microbiol. 12, 158(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Santiago A et al. Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol. 14, 112 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wild CP Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 14, 1847–1850 (2005). [DOI] [PubMed] [Google Scholar]; A review of the important impact of the exposome on biomarker development.
- 117.Spitz MR et al. An expanded risk prediction model for lung cancer. Cancer Prev. Res. (Phila.) 1, 250–254 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Spitz MR et al. A risk model for prediction of lung cancer. J. Nati Cancer inst 99, 715–726 (2007). [DOI] [PubMed] [Google Scholar]
- 119.Wender R et al. American Cancer Society lung cancer screening guidelines. CA Cancer J. Clin 63, 107–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Khariwala SS et al. Self-reported tobacco use does not correlate with carcinogen exposure in smokers with head and neck cancer. Laryngoscope 125, 1844–1848 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fagan P et al. Biomarkers of tobacco smoke exposure in racial/ethnic groups at high risk for lung cancer. Am. J. Public Health 105, 1237–1245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hecht SS Lung carcinogenesis by tobacco smoke. Int. J. Cancer 131, 2724–2732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Imielinski M et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Ceil 150, 1107–1120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gibbons DL, Byers LA & Kurie JM Smoking, p53 mutation, and lung cancer. Mol. Cancer Res 12, 3–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takahashi T et al. p53: a frequent target for genetic abnormalities in lung cancer. Science 246, 491–494 (1989). [DOI] [PubMed] [Google Scholar]
- 126.Dobbin KK et al. Interlaboratory comparability study of cancer gene expression analysis using oligonucleotide microarrays. Clin. Cancer Res 11, 565–572 (2005). [PubMed] [Google Scholar]
- 127.Florkowski CM Sensitivity, specificity, receiver-operating characteristic (ROC) curves and likelihood ratios: communicating the performance of diagnostic tests. Clin. Biochem. Rev 29 (Suppl. 1), S83–S87 (2008). [PMC free article] [PubMed] [Google Scholar]
- 128.Pagano M & Gauvreau K Principles of Biostatistics (ed. Crockett C) (Brooks/Cole, 2000). [Google Scholar]
- 129.Polley MY et al. Statistical and practical considerations for clinical evaluation of predictive biomarkers. J. Natl Cancer Inst 105, 1677–1683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; A summary of the statistical challenges and considerations for biomarker development.
- 130.Rothman KJ Epidemiology: An introduction (Oxford Univ. Press, 2012). [Google Scholar]
- 131.Schork NJ Personalized medicine: time for one-person trials. Nature 520, 609–611 (2015). [DOI] [PubMed] [Google Scholar]
- 132.Riely GJ et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin. Cancer Res 14, 5731–5734 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yuan Y et al. Assessing the clinical utility of cancer genomic and proteomic data across tumor types. Nat Biotechnoi 32, 644–652 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saito M et al. The association of microRNA expression with prognosis and progression in early-stage, non-small cell lung adenocarcinoma: a retrospective analysis of three cohorts. Clin. Cancer Res 17, 1875–1882(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Robles AI et al. An integrated prognostic classifier for stage 1 lung adenocarcinoma based on mRNA, microRNA and DNA methylation biomarkers. J. Thorac. Oncol 10, 1037–1048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; A report on a validated, multiple ‘omics’ biomarker panel for lung adenocarcinoma.
- 136.Liu CY et al. Genome-wide gene-asbestos exposure interaction association study identifies a common susceptibility variant on 22ql 3.31 associated with lung cancer risk. Cancer Epidemiol. Biomarkers Prev 24, 1564–1573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT01248247?term-NCT01248247&rank=1 (2016). [DOI] [PubMed]; An example of a precision medicine clinical trial.
- 138.US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/Ct2/show/NCT01506045?term-NCT01506045&rank=1 (2016). [DOI] [PubMed]
- 139.Glass K, Huttenhower C, Quackenbush J & Yuan GC Passing messages between biological networks to refine predicted interactions. PLoS ONE 8, e64832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Le Cao KA, Boitard S & Besse P Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics 12, 253 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liquet B, Le Cao KA, Hocini H & Thiebaut R A novel approach for biomarker selection and the integration of repeated measures experiments from two assays. BMC Bioinformatics 13, 325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Valente AX & Cusick ME Yeast protein interactome topology provides framework for coordinated-functionality. Nucleic Acids Res. 34, 2812–2819 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang B et al. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 11, 333–337(2014). [DOI] [PubMed] [Google Scholar]
- 144.Li L et al. Integrated omic analysis of lung cancer reveals metabolism proteome signatures with prognostic impact. Nat. Commun 5, 5469 (2014). [DOI] [PubMed] [Google Scholar]
- 145.Kim SC et al. A high-dimensional, deep-sequencing study of lung adenocarcinoma in female never-smokers. PLoS ONE 8, e55596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.International Cancer Genome Consortium. International network of cancer genome projects. Nature 464, 993–998 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Erler JT & Unding R Network-based drugs and biomarkers. J. Pathol 220, 290–296 (2010). [DOI] [PubMed] [Google Scholar]
- 148.Greene CS et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet 47, 569–576 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Taylor IW et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat. Biotechnoi 27, 199–204 (2009). [DOI] [PubMed] [Google Scholar]
- 150.Chin CD, Linder V & Sia SK Commercialization of microfluidic point-of-care diagnostic devices. Lab. Chip 12, 2118–2134 (2012). [DOI] [PubMed] [Google Scholar]
- 151.Day N et al. EPIC-Norfolk: study design and characteristics of the cohort. European Prospective Investigation of Cancer. Br. J. Cancer 80 (Suppl. 1), 95–103 (1999). [PubMed] [Google Scholar]
- 152.Women’s Health Initiative Study Group. Design of the Women’s Health Initiative clinical trial and observational study. Control. Clin. Trials 19, 61–109 (1998). [DOI] [PubMed] [Google Scholar]
- 153.McCarty CA et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genom 4, 13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Altman DG, McShane LM, Sauerbrei W & Taube SE Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. PLoS Med. 9, el 001216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Altman DG, McShane LM, Sauerbrei W & Taube SE Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med. 10, 51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 154 and 155 provide guidelines on how tumour biomarker studies should be reported to improve the field.
- 156.McShane LM et al. Reporting recommendations for tumor marker prognostic studies (REMARK). Exp. Oncol 28, 99–105 (2006). [PubMed] [Google Scholar]
- 157.McShane LM et al. Reporting recommendations for tumor marker prognostic studies (REMARK). Nat. Clin. Pract. UroL 2, 416–422 (2005). [PubMed] [Google Scholar]
- 158.McShane LM et al. Reporting recommendations for tumor marker prognostic studies. J. Clin. Oncol 23, 9067–9072 (2005). [DOI] [PubMed] [Google Scholar]
- 159.McShane LM et al. Reporting recommendations for tumor marker prognostic studies (REMARK). Nat. Clin. Pract Oncol 2, 416–422 (2005). [PubMed] [Google Scholar]
- 160.McShane LM et al. Reporting recommendations for tumour marker prognostic studies (REMARK). Br.J. Cancer 93, 387–391 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.McShane LM et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J. Natl Cancer Inst 97, 1180–1184 (2005). [DOI] [PubMed] [Google Scholar]
- 162.McShane LM et al. Reporting recommendations for tumour marker prognostic studies (REMARK). Eur. J. Cancer 41,1690–1696 (2005). [DOI] [PubMed] [Google Scholar]
- 163.Srivastava S The early detection research network: 10-year outlook. Clin. Chem 59, 60–67 (2013). [DOI] [PubMed] [Google Scholar]
- 164.McRonald FE et al. The UK Lung Screen (UKLS): demographic profile of first 88,897 approaches provides recommendations for population screening. Cancer Prev. Res. (Phila.) 7, 362–371 (2014). [DOI] [PubMed] [Google Scholar]
- 165.National Lung Screening Trial Research Team. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med 365, 395–409(2011). [DOI] [PMC free article] [PubMed] [Google Scholar]; Reports on findings that support the use of LDCT to screen for lung cancer.
- 166.Schmidt C Lung cancer screening poised to expand. J. Natl Cancer Inst 107, djvll4 (2015). [DOI] [PubMed] [Google Scholar]
- 167.Ma J, Ward EM, Smith R & Jemal A Annual number of lung cancer deaths potentially avertable by screening in the United States. Cancer 119, 1381–1385 (2013). [DOI] [PubMed] [Google Scholar]
- 168.Pinsky PF & Berg CD Applying the National Lung Screening Trial eligibility criteria to the US population: what percent of the population and of incident lung cancers would be covered? J. Med. Screen 19, 154–156 (2012). [DOI] [PubMed] [Google Scholar]
- 169.Patz EF Jr et al. Lung cancer incidence and mortality in National Lung Screening Trial participants who underwent low-dose CT prevalence screening: a retrospective cohort analysis of a randomised, multicentre, diagnostic screening trial. Lancet Oncol. http://dx.doi.org/10,1016/S1470-2045(15)00621-X (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lee HY et al. Role of CT and PET imaging in predicting tumor recurrence and survival in patients with lung adenocarcinoma: a comparison with the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification of lung adenocarcinoma. J. Thorne. Oncol 10, 1785–1794 (2015). [DOI] [PubMed] [Google Scholar]
- 171.Ruparel M et al. Pulmonary nodules and CT screening: the past, present and future. Thorax 71, 367–375(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Elmore JG et al. Ten-year risk of false positive screening mammograms and clinical breast examinations. N. Engl. J. Med 338, 1089–1096 (1998). [DOI] [PubMed] [Google Scholar]
- 173.Ahmad A & Gadgeel SM (eds) Lung Cancer and Personalized Medicine (Springer, 2016). [PubMed] [Google Scholar]
- 174.Alberg AJ & Samet JM Epidemiology of lung cancer. Chest 123, 21S–49S (2003). [DOI] [PubMed] [Google Scholar]