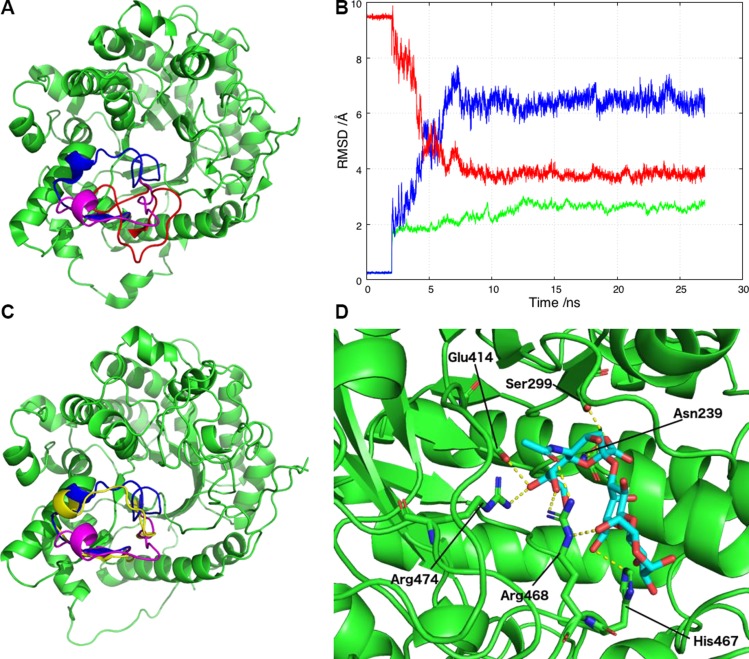
Figure 4.
αKL1 binds to α2-3-sialyllactose and allows the open conformation of the β6α6 loop to relocate to one closer to the X-ray structure during the MD simulation. A) Three β6α6 loop configurations in αKL1: the X-ray configuration (blue), the revised conformation (magenta), and the previous homology model conformation (red). B) RMSD plot against the initial starting model for the αKL1 non-β6α6 loop backbone atoms (green) and the β6α6 loop backbone atoms (blue) during the simulations. Superimposed (red) is the RMSD of the β6α6 loop backbone atoms against the X-ray structure. C) During the simulations, the β6α6 loop changes conformation from the starting homology model open conformation used for docking (magenta) to the final conformation (yellow) that is closer to the X-ray conformation (blue). D) A snapshot from the simulations of αKL1 (light green) and α2-3-sialyllactose (cyan with O atoms in red) showing their potential hydrogen bonds. The sialic acid carboxylate group moves closer to the Asn239 and Ser299 sidechains to accept a hydrogen atom from each. The sialic acid hydroxyl group serves as a hydrogen bond donor to Glu414 and a hydrogen bond receptor from Arg474. Arg468 participates in multiple hydrogen bonds with the glucose and the sialic acid, whereas His467 forms a hydrogen bond with galactose O6.