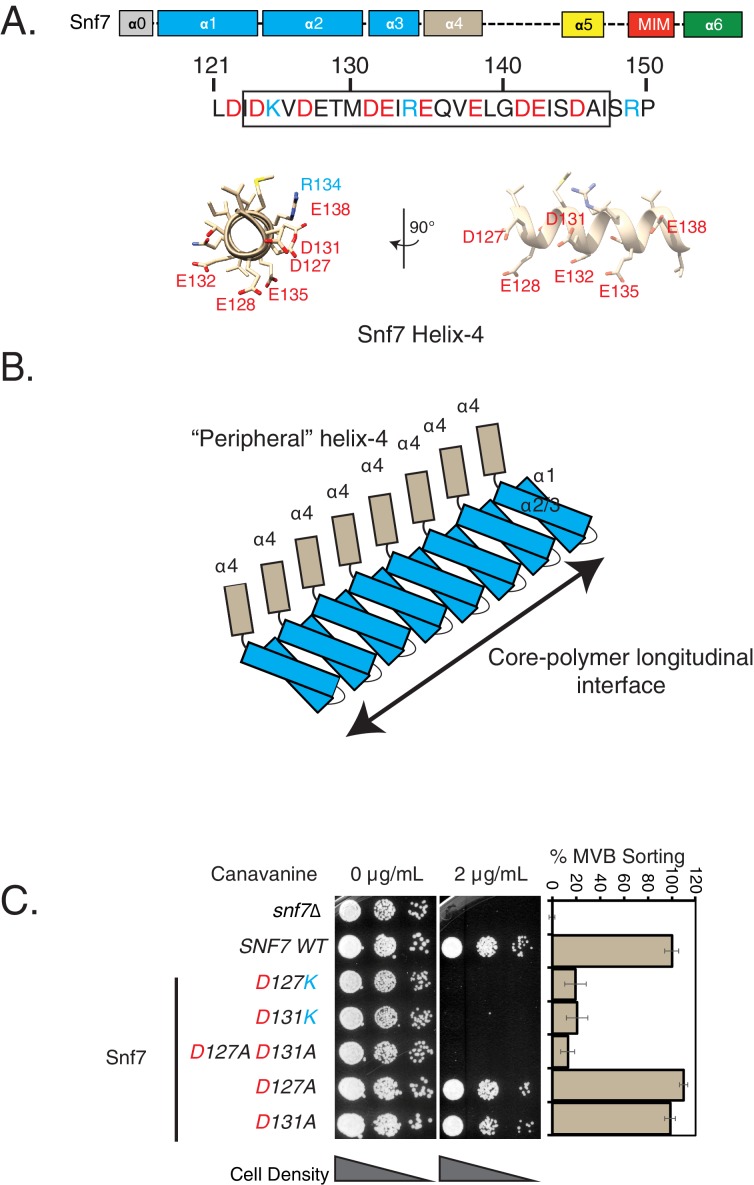
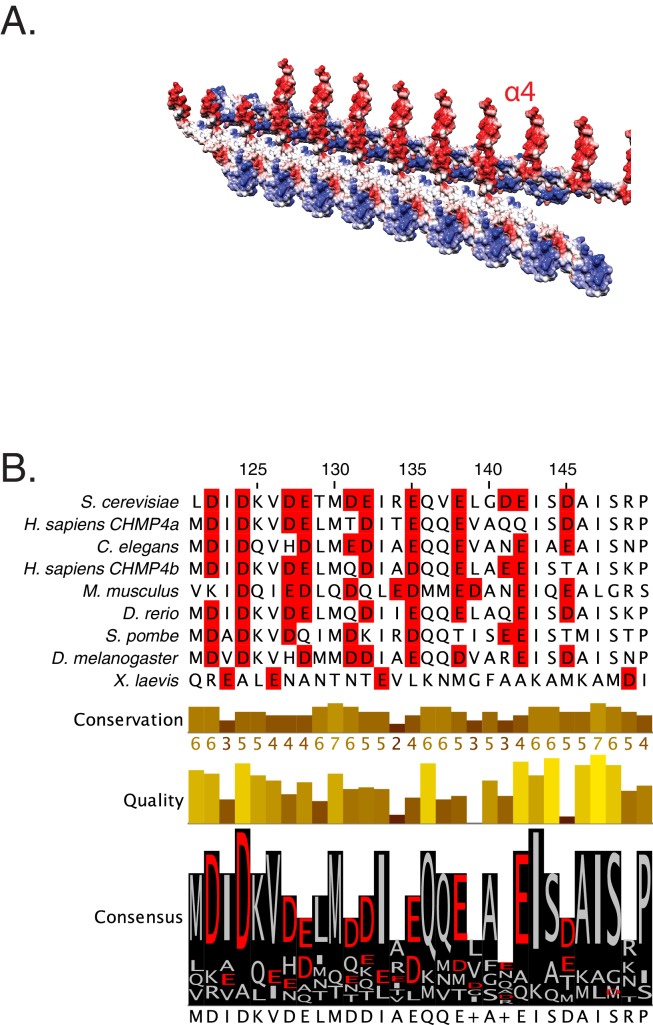
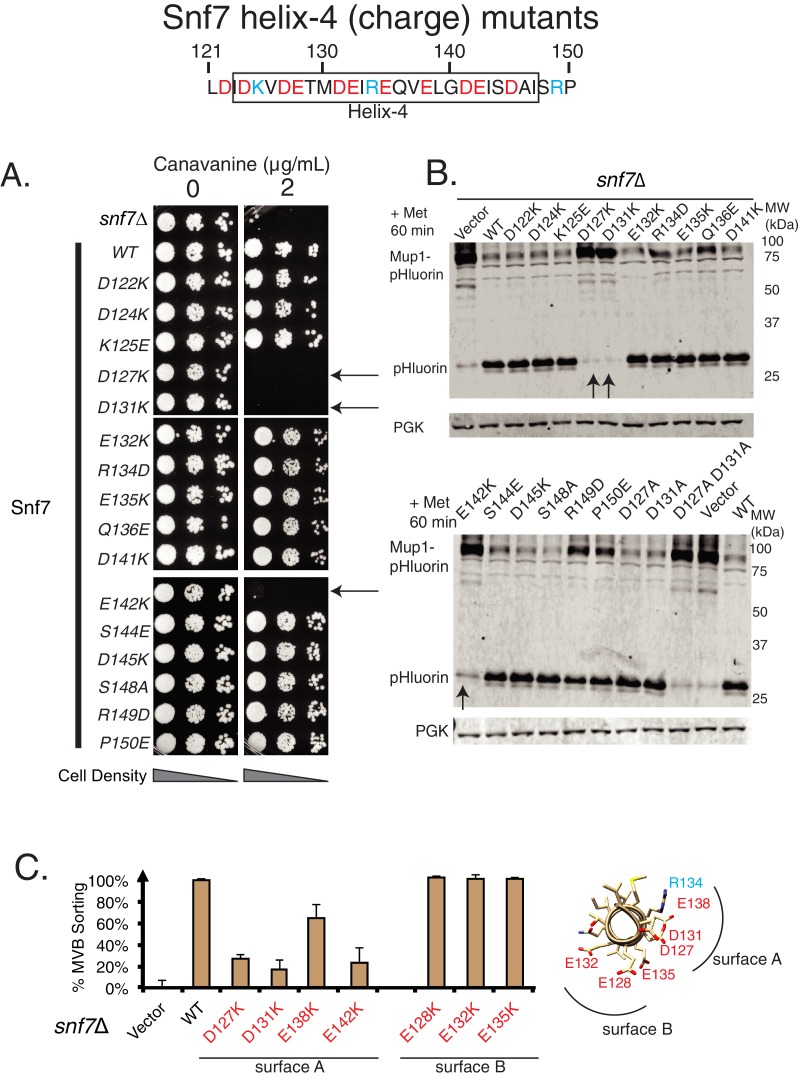
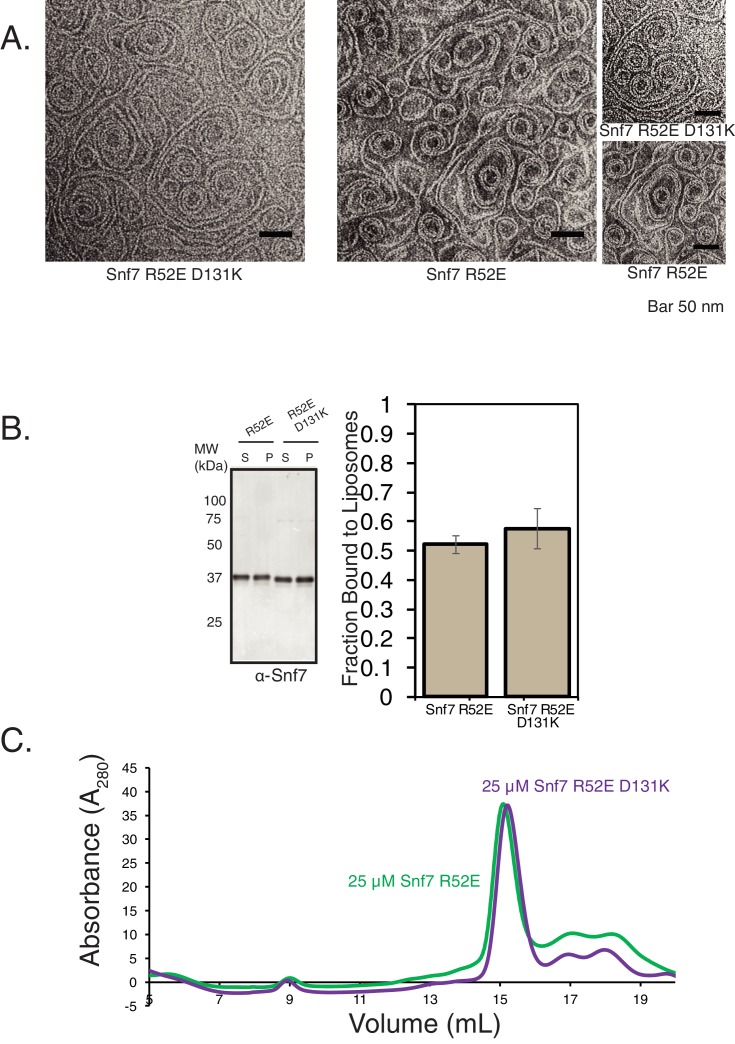
Figure 1. Mutations in helix-4 region of Snf7 induce cargo-sorting defect.
(A) Domain organization of Snf7, depicting the different helices (top). Bottom figure shows the sequence of helix-4, with the predicted helical motif highlighted with a box. Acidic residues are denoted in red, while cyan residues are basic amino acids. Bottom – structure of the helix-4 (from PDB 5fd7) in two orientations, highlighting the acidic residues on one surface. (B) Cartoon model of the polymeric arrangement of Snf7 in its linear form observed in the crystal lattice. (C) Canavanine sensitivity and Mup1-pHluorin flow-cytometry data (right) showing cargo-sorting/endocytosis defects of the helix-4 mutants of Snf7. Mup1-pHluorin data were collected 90 min after methionine addition. Error bars represent standard deviation from 3 to 7 independent experiments.