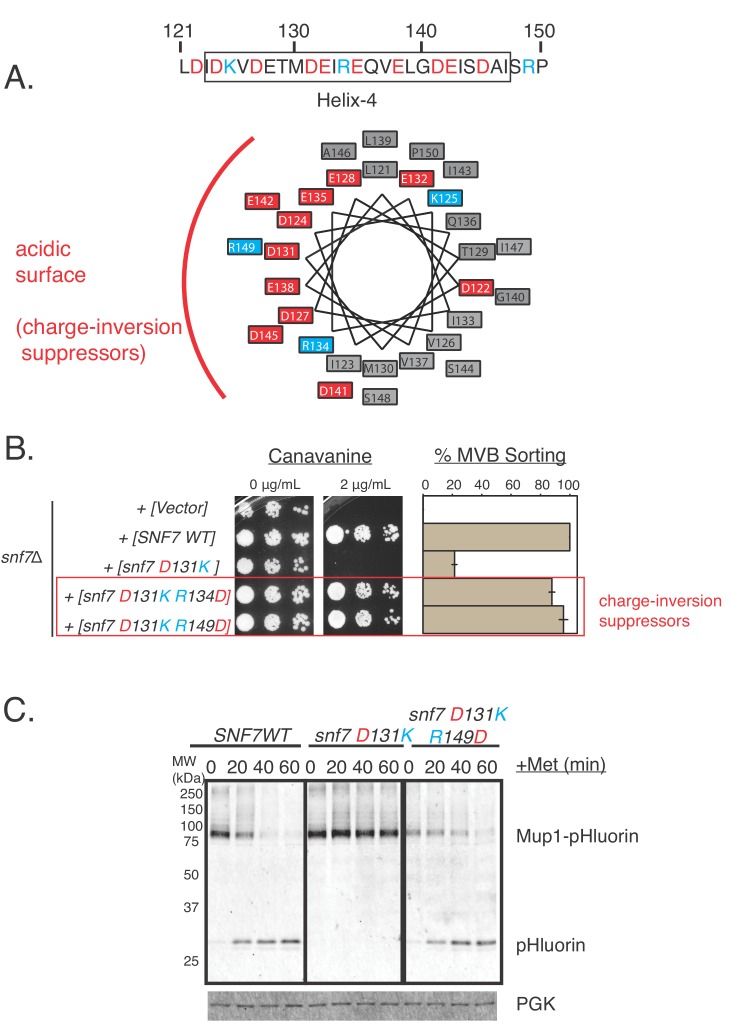
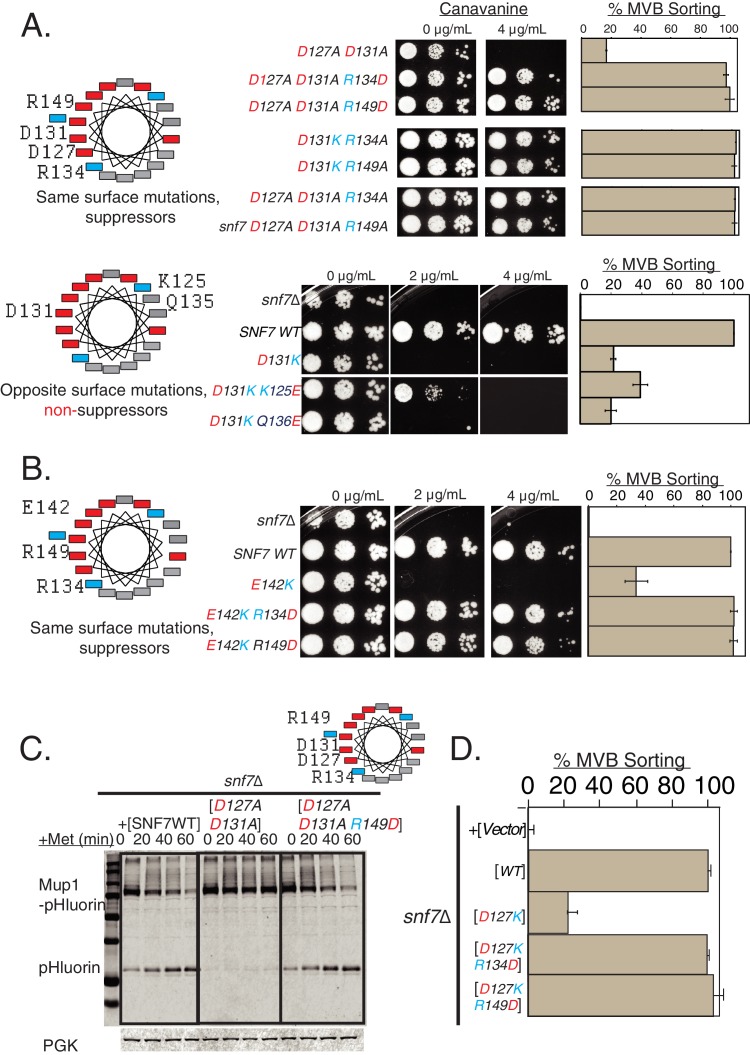
(A) Helix-wheel on the left indicate the positions of the mutations analyzed in this figure. Canavanine sensitivity or Mup1-pHluorin assays with opposite charge mutations in helix-4. With D127A D131A mutations including the R134D or R149D, the endocytosis defect is restored. Mutating R134 or R149 to alanine also suppress the defect of D131K, and also suppress the mutant D127A D131A. Bottom-panel depict mutations in opposite surface of helix-4, which do not suppress the defects observed in endocytosis. Error bars represent standard deviation from three independent experiments. (B) Similar Can1 and Mup1 assays with mutations at the E142 location of Snf7. The same surface charge-rebalancing mutations (E142K R134D or E142K R149D) are completely functional in cargo-sorting assays. Error bars represent standard deviation from three independent experiments. (C) Mup1-pHluorin degradation over time after addition of methionine with mutants snf7 D127A D131A and snf7 D127A D131A R149D, blotting for pHluorin. Adding a Glu in place of Arg to helix-4 rescues the defect of the double mutation D127A D131A. (D) Quantification of the Mup1-pHluorin assays with mutations at the D127 position of Snf7, whose defect is rescued by charge-balancing mutations in the helix. Error bars represent standard deviation from three independent experiments.