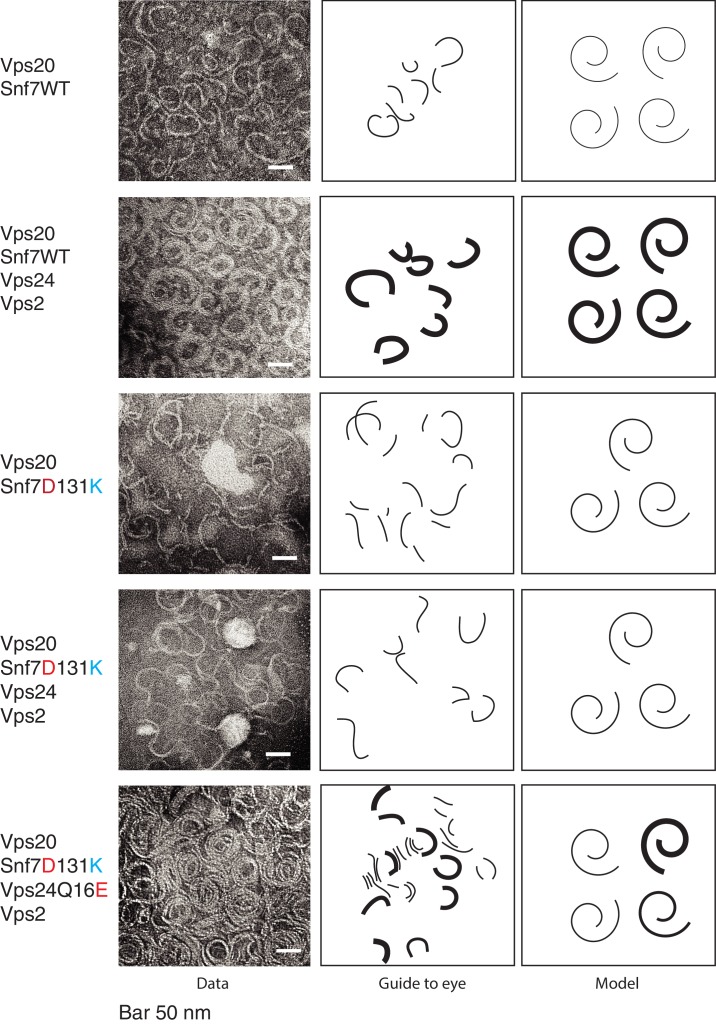
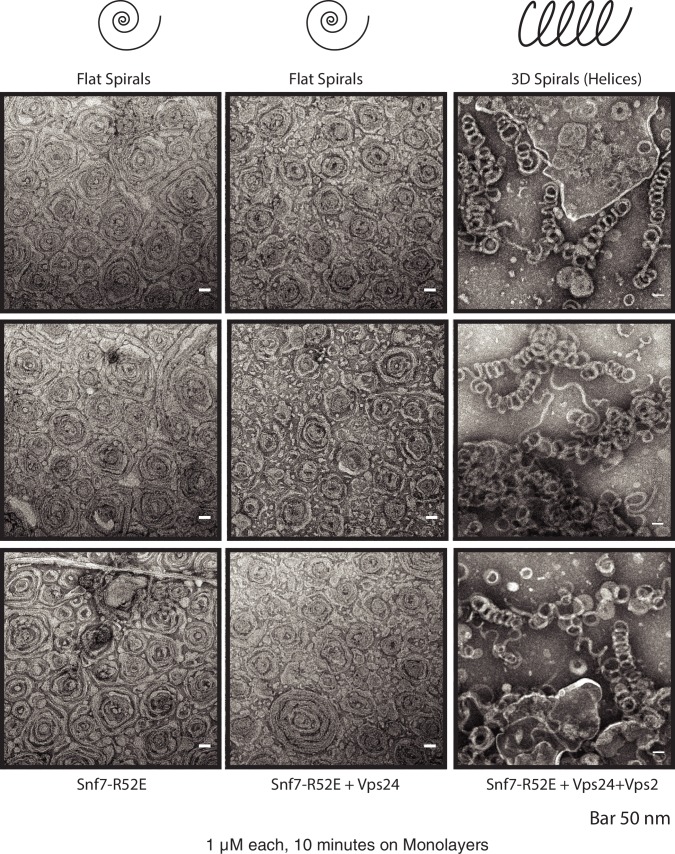
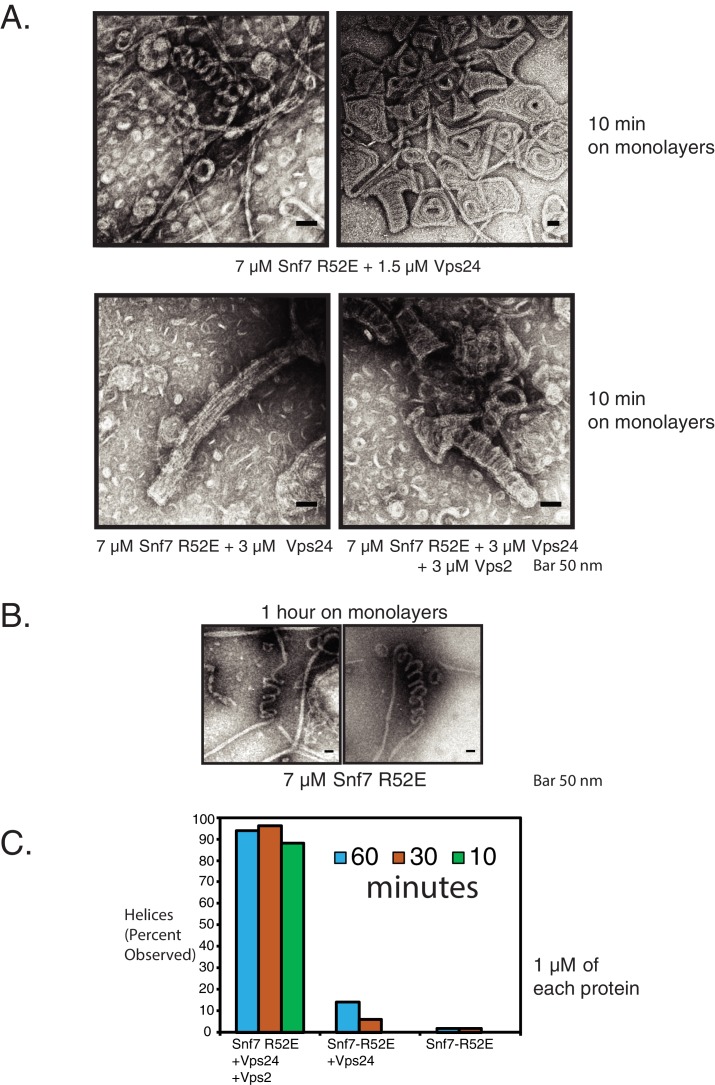
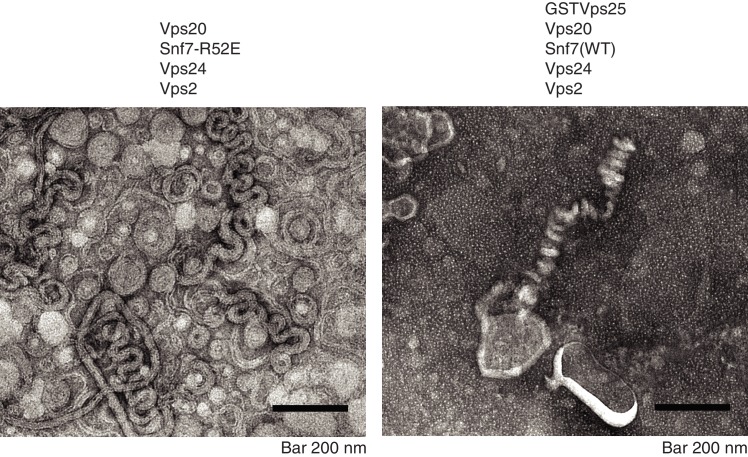
Figure 5. Vps24/Vps2 form lateral copolymer with Snf7.
(A) Electron microscopy analysis of 10 μM Vps20, 1 μM Snf7 or (Snf7 D13K), 1 μM Vps24 (or Vps24 Q16E) and 1 μM Vps2. Experiments were done on lipid monolayers, with incubation of proteins for 1 hr. Painted lines on the middle-panel are guide to the eye to demonstrate differences in the thickness of the copolymers. The right panel shows hypothetical model of the structures of the polymers made from the different proteins.