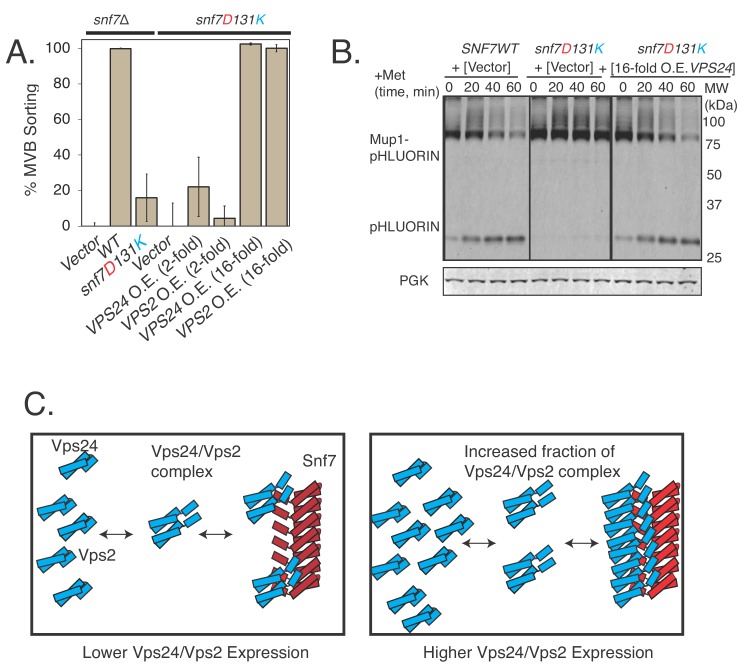
Figure 6. Vps24 and Vps2 cooperatively bind Snf7, as over-expressing them suppresses the defects of Snf7 helix-4 mutants.
(A) Mup1-pHluorin endocytosis data after 90 min of methionine addition, showing overexpression of Vps24 or Vps2 rescuing the defect of the helix-4 mutant D131K. 2-fold overexpression (O.E.) corresponds to expressing either Vps24 or Vps2 with a CEN plasmid, while ~16 fold represents experiments performed with these genes on a CMV promoter with a tet-off operator. Error bars represent standard deviation from three to five independent experiments. (B).Overexpression of Vps24 (on a plasmid with a tet-off operator), rescues Mup1-pHluorin degradation defect of the D131K mutant, as shown immunoblotting of pHluorin over 60 min of methionine addition. (C).Model showing the hypothesized recruitment of Vps24/Vps2 in helix-4 mutant under normal (left) or under overexpressed (right) Vps24/Vps2 conditions.