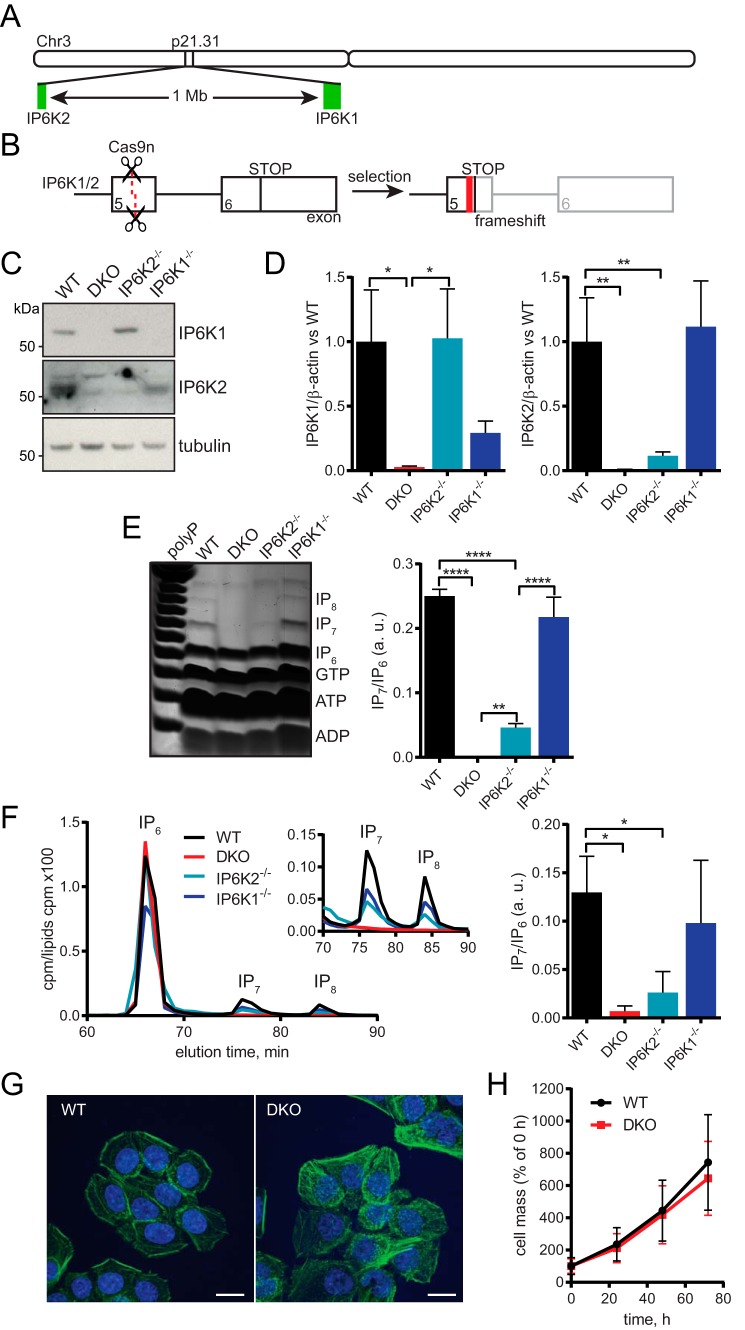
Figure 1.
Generation and characterization of IP6K knockout HCT116 cells. A, schematic showing the localization of human IP6K1 and IP6K2 on chromosome 3, to scale. In mice, these genes are separated by 0.7 Mb on chromosome 9 at 9 F1 and 9 F2, respectively. B, schematic for creation of IP6K1/2 knockout human cells using CRISPR. Guide RNAs were designed to target exon 5 of IP6K1 and IP6K2, using nickase Cas9n D10A. Successfully mutated clones contained indels that caused frameshift and premature stop codons. C, Western blotting for IP6K1 and IP6K2 showing their loss in KO cells. Tubulin is shown as loading control. D, RT-qPCR analysis of IP6K1 (left) and IP6K2 (right) mRNA transcripts in KO cells, normalized to β-actin. We were unable to detect any IP6K3 protein or mRNA. E, titanium dioxide-purified perchloric acid cell extracts resolved by 35% PAGE and stained with toluidine blue, with densitometric analysis (right). Identity of bands determined by migration compared with previous analysis of standards. Extracts equivalent to 16 mg of protein were loaded. Synthetic polyP used as a ladder. F, SAX-HPLC of myo-[3H]inositol–labeled cells. Cells were labeled for 5 days in inositol-free DMEM. Inset shows a close up of IP7 and IP8. Ratio of the IP7/IP6 peaks is shown (right). G, maximum projection images of FITC-phalloidin (green) stained fixed cells. Nuclei stained with Hoechst (blue). Scale bars, 15 μm. H, SRB cell growth assay. Data show mean ± S.D. from 11 experiments. Bar charts in B–D show mean ± S.D. from 3 experiments. HPLC traces in D and images in E are representative of experiments performed 3 times. *, p < 0.05; **, p < 0.01; ****, p < 0.0001, ANOVA with Tukey post test.