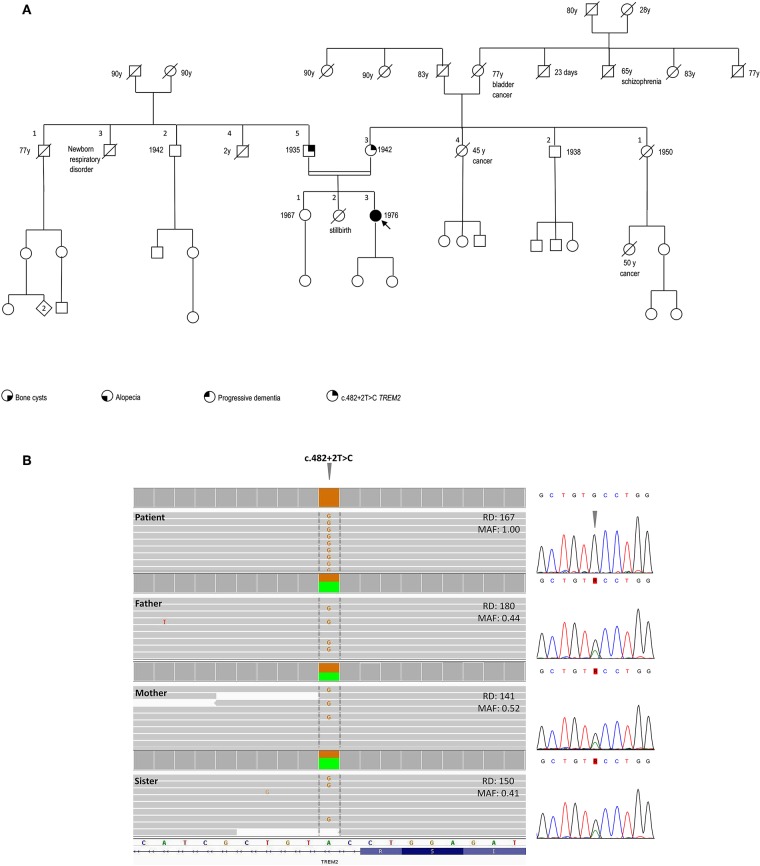
Figure 2.
Pedigree and genetic findings of the index case. (A) Pedigree of the family and recorded clinical phenotypes. (B) IGV (Integrative Genomics View) visualization (right panel) of the chr6:41,127,528A>G (GRCh37/hg19) splice-site variant c.482+2T>C in patient (homozygous) and relatives (heterozygous). The variant, disrupting the donor splice site of TREM2 intron 3, is annotated in both ClinVar (#RCV000050138.1) and HGMD (#CS022111) as a disease-causing/likely pathogenic and has an extremely rare allele frequency in population databases (gnomAD: 0.0008126%; ExAC: 0.00165%), with only two reported heterozygous individuals in the European Non-Finnish population and no homozygotes. The variant is included in one of the numerous regions of homozygosity (chr6:24,747,411-42,365,687). Read depths (DP) and Mutant Allele Fraction (MAF) of the variant are shown for each member of the family. Sanger sequencing confirmation of the TREM2 variant is shown in the left panel.