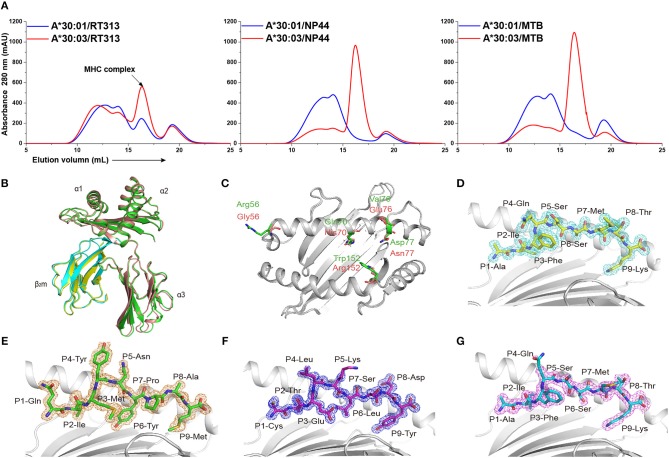
Figure 1.
The binding capabilities and overall structures of HLA-A*30:01 and HLA-A*30:03. (A) Binding of RT313, NP44, and MTB to HLA-A*30:01 (blue) or HLA-A*30:03 (red) was elucidated by in vitro refolding assays. Peptides presented by HLA-A*30:01 or HLA-A*30:03 help their H chain and human β2 microglobulin to refold in vitro. After correct refolding, the high absorbance peaks of the HLAs with the expected molecular mass of 45 KD were eluted at an estimated volume of 16 mL on a SuperdexTM 200 10/300 GL column. (B) Structure alignment of HLA-A*30:01 (green) and HLA-A*30:03 (salmon) showed a similar overall conformation between the two alleles. (C) The amino acids that differed between HLA-A*30:01 (green) and HLA-A*30:03 (salmon) are shown as sticks. The authentic conformations of peptides RT313 (D) in the HLA-A*30:01 grooves and MTB (E), NP44 (F), and RT313 (G) in the HLA-A*30:03 grooves are shown in the 2Fo-Fc electron density maps contoured at 1.0 s. The peptide from one molecule in the asymmetric unit of each of the two structures is shown as a representative example. The residues in the peptides are labeled as individual amino acids with their position numbers.