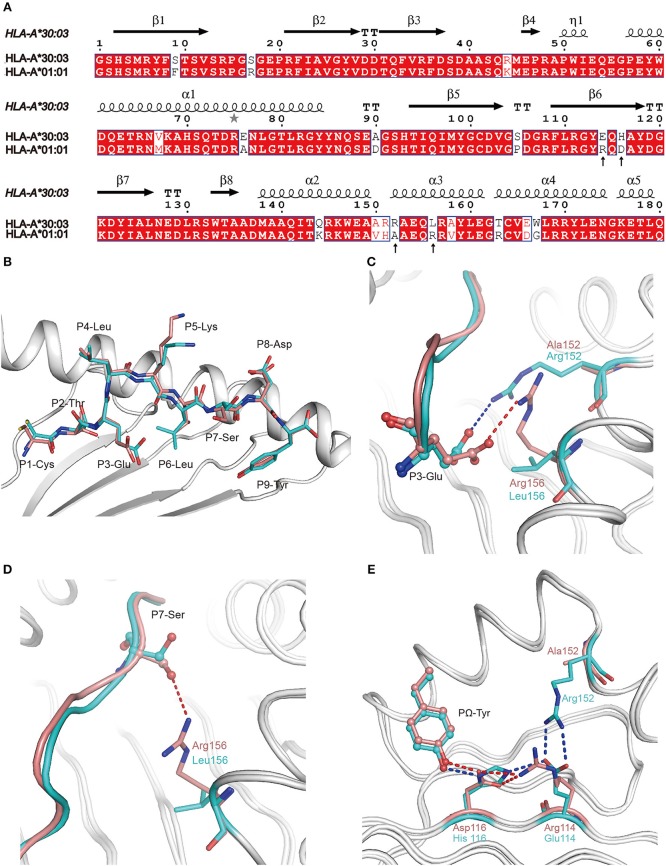
Figure 4.
The contribution of polymorphic amino acids to the different construction of NP44 presented by HLA-A*30:03 and HLA-A*01:01 and to the recognition of TCRs. (A) Structure-based sequence alignment of HLA-A*30:03 and HLA-A*01:01, addressing the α1 and α2 domains. Cylinders indicate α-helices and black arrows indicate β-strands. Amino acids highlighted in red are completely conserved and those in blue boxes are highly (>80%) conserved. Sequence alignment was generated with Clustal X (33) and ESPript (34). (B) Superposition of NP44 presented by HLA-A*30:03 and HLA-A*01:01. The overall conformations of NP44 in HLA-A*30:03 and HLA-A*01:01 were very similar. However, P3-Glu, P5-Lys, and P7-Ser showed slightly different conformations in these two structures. (C) The different conformations of P3-Glu of NP44 presented by HLA-A*30:03 (cyan) and HLA-A*01:01 (lemon). The hydrogen bonds between P3-Glu and residues in HLA-A*30:03 are represented as blue dashed lines and those with residues in HLA-A*01:01 are represented as red dashed lines. (D) The different conformations of P7-Ser of NP44 presented by HLA-A*30:03 (cyan) and HLA-A*01:01 (lemon). The hydrogen bonds between P7-Ser and Arg156 in HLA-A*01:01 are represented as red dashed lines While, Leu156 in HLA-A*30:03 could not form hydrogen bonds with P7-Ser. (E) Different hydrogen bond networks of PΩ-Tyr of NP44 with residues 114 and 116 of HLA-A*30:03 and HLA-A*01:01. Hydrogen bonds are represented as blue dashed lines in HLA-A*30:03/NP44 and as red dashed lines in HLA-A*01:01/NP44.