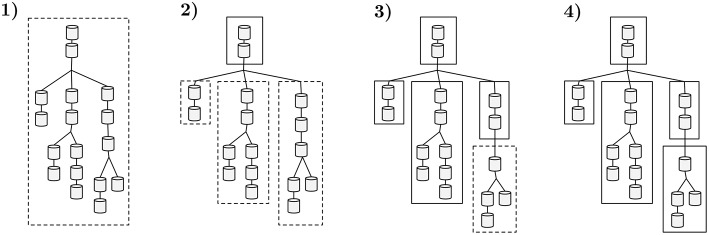
Figure 5.
A sample workflow of the algorithm that decomposes neuron morphologies into a tree of subtrees. Dashed contours display groups of compartments being tested against the maximum computational complexity threshold. Straight contours represent a set of compartments with a total computational complexity below the threshold, clustered into a subtree. (1) The total runtime of the initial tree is computed and compared with the complexity threshold. (2) The previous cluster exceeds the allowed computational complexity threshold. The following sequence of non-bifurcated compartments that yields smaller complexity (top two compartments) is clustered into a subtree. The connecting three branches are tested. (3) Both left and center sections are benchmarked and their execution time is below the threshold, thus becoming two independent subtrees. The same threshold test for the right region fails. The first two compartments in the right region yield less runtime than the complexity threshold and are clustered into a subtree. (4) The remaining compartments on the right branch of the tree comply with the complexity test and are clustered, leading to the final data representation.