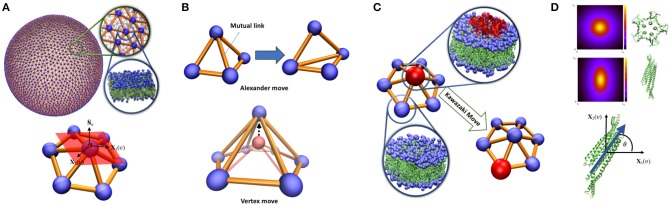
Figure 1.
(A) Triangulated surface representation of a vesicle. Each vertex represents a segment of a bilayer containing hundreds of lipids. On each vertex, surface normal (), principal directions X1(υ), X2(υ) and their associated principal curvature (c1υ, c2υ) can be determined. (B) Top: Alexander move, mutual link between neighboring triangles is flipped and two new triangles are generated. Bottom: vertex move, a chosen vertex (red bead) is moved in a random direction (C) Proteins are modeled as an inclusion that can lay on a vertex and can jump to the neighboring vertex via Kawazaki moves. (D) Vertex based model of curvature map induced by proteins with lateral symmetry higher than 2 (top) and π-symmetric proteins (middle). The inclusion associated with these proteins should have a character of a two-dimensional vector in the plane of a vertex (bottom). The angle between the protein direction and the membrane main principal direction.