Figure 1.
Nocturnin Localizes to Both the Mitochondria and Cytosol via the Use of Alternative Translation Initiation Sites
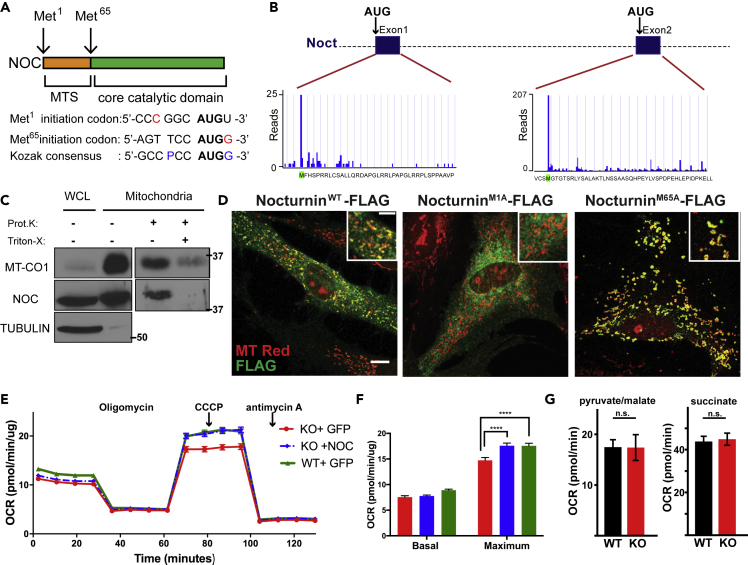
(A) Nocturnin contains two potential initiation codons (Met1 and Met65), separated by a mitochondrial targeting sequence. Nucleotides in red show the critical components of the Kozak sequence: a purine (A or G) at position −3 and a G following the AUG start codon.
(B) Quantitative translation initiation sequencing (QTI-seq) analysis of human Nocturnin; graphs and data reproduced from GWIPS-viz database (Michel et al., 2014).
(C) Proteinase K (Prot.K) protection assay of the mitochondrial fraction from HEK293 cells. Mitochondrial fractions were treated with Proteinase K (100 μg/mL) or Triton X-100 (1%), and samples were blotted for MT-CO1 (mitochondrial) and TUBULIN (cytoplasmic) as controls. WCL, whole-cell lysate. Molecular weight markers are indicated in kilodaltons.
(D) Nocturnin−/− MEFs expressing exogenous versions of Nocturnin were stained with MitoTracker (red; mitochondria), FLAG antibody (green; NOCTURNIN). Scale bar, 21μM.
(E) Oxygen consumption rates (normalized to protein concentration) in cells of the indicated genotype. Oligomycin, CCCP (carbonyl cyanide m-chlorophenylhydrazone), and antimycin A were injected at the indicated times.
(F) Bar graph showing averages for basal and maximum respiration. A representative experiment from three independent experiments is shown.
(G) State 4 respiration from permeabilized cells of the indicated genotype in response to complex I (pyruvate/malate) or complex II (succinate) substrates.
N = 16–23 per group, ****p < 0.0001, p values are calculated by two-way ANOVA followed by Tukey's post-hoc test. Data are represented as mean ± SEM; n.s., not significant. See also Figure S1.