Figure 3.
Altered Expression of Mitochondrial Metabolic Genes in Nocturnin−/− BAT Following Cold Exposure
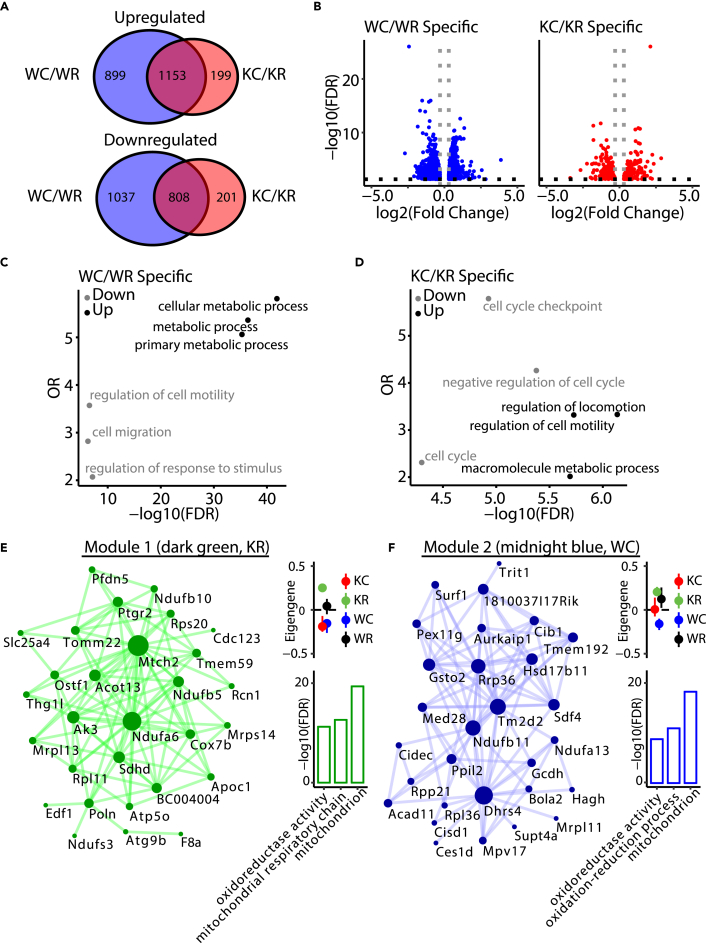
(A) Venn diagrams of common and unique genes that are differentially expressed following 4-h cold exposure in wild-type (WC/WR) and Nocturnin−/− (KC/KR) BAT. Top: Venn diagram of upregulated genes. Bottom: Venn diagram, downregulated genes. Blue, WT; red, Nocturnin−/−. WR, wild-type room temperature; WC, wild-type cold; KR, Nocturnin−/− room temperature; KC, Nocturnin−/− cold.
(B) Volcano plot for uniquely changing genes in WT and Nocturnin−/− BAT following 4-h cold exposure. y axis represents –log10(FDR). x axis represent log2(fold change) from differential expression analysis statistics.
(C and D) Functional enrichment analyses of uniquely differentially expressed genes in (C) WT (# of genes = 1,935) and (D) Nocturnin−/− (# of genes = 399). Downregulated pathways are shown in gray; upregulated pathways are shown in black. y axis represents odds ratios (OR). x axis represents –log10(FDR). The top three most significant categories are shown.
(E and F) Visualization of the top 100 connections ranked by weighted topological overlap values for module 1 (dark green, KR correlated) and module 2 (midnight blue, WC correlated). Node size corresponds to the number of edges (degree). Side dot plots with standard errors demonstrate the association of the modules detected with genotype and temperature. Standard errors are calculated based on the eigengene across samples. Dots represent the mean eigengene for that module. Side bar plots show the top three gene ontology groups of the module based on –log10(FDR). FDR, false discovery rate.