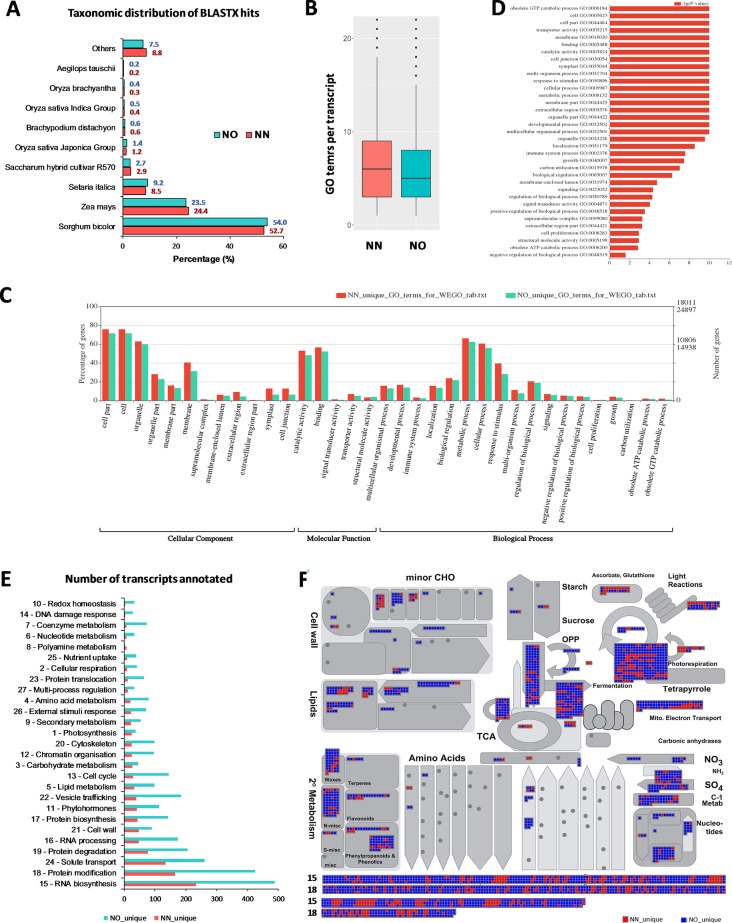
Figure 5.
Functional annotation of transcript isoforms from the two datasets. (A) Taxonomic distribution of BLATX hits of the two datasets. (B) GO terms per transcripts. (C) Significantly different GO terms between the two datasets. (D) Significantly different GO terms with highest log10(p value) identified from the two datasets. (E) Unique bins annotated using Arabidopsis genes that matched the unique fractions of the NN and NO datasets. (F) MapMan functional bins identified from the two datasets, red heatmap represents transcripts from the NN dataset, while blue represents those from the NO dataset. Two bins (15 and 18) were added to the bottom of the panel listing all matched genes from two datasets. Each heatmap represents one annotated transcript. NN, sugarcane non-normalized PacBio Iso-Seq isoforms; NO, sugarcane normalized PacBio Iso-Seq isoforms.