Abstract
Bacteria that oxidize methane to methanol are central to mitigating emissions of methane, a potent greenhouse gas. The nature of the copper active site in the primary metabolic enzyme of these bacteria, particulate methane monooxygenase (pMMO), has been controversial owing to seemingly contradictory biochemical, spectroscopic, and crystallographic results. We present biochemical and electron paramagnetic resonance spectroscopic characterization most consistent with two monocopper sites within pMMO: one in the soluble PmoB subunit at the previously assigned active site (CuB) and one ~2 nanometers away in the membrane-bound PmoC subunit (CuC). On the basis of these results, we propose that a monocopper site is able to catalyze methane oxidation in pMMO.
Methane is both a potent greenhouse gas and a readily available energy source (1–3). Methanotrophic bacteria use enzymes called methane monooxygenases (MMOs) to activate dioxygen and break a 105 kcal/mol C–H bond in methane to produce methanol at ambient pressure and temperature(1). By contrast, current industrial catalysis processes for this reaction require tremendous pressure and high temperature (>1000 K). Understanding how enzymes catalyze this reaction is critical to the development of catalysts that function at moderate conditions (4–8).
The most common MMO is the membrane-bound, copper-dependent particulate enzyme (pMMO) (9). Multiple pMMO crystal structures reveal a trimeric assembly of protomers, each comprising two predominantly transmembrane subunits (PmoA and PmoC) and one transmembrane subunit with a large periplasmic domain (PmoB) (Fig. 1A) (10–13). Three copper-binding sites have been detected in the pMMO structures, (i) A monocopper site, denoted as the bis-His site, is ligated by His48 and His72 (fig. S1). However, His48 is not conserved, and this site is observed only in the Methylococcus capsidatus (Bath) pMMO structure (10), so it is not believed to play a critical role in catalysis (14). (ii) All structures contain a site denoted CuB, in which copper is coordinated by the amino-terminal histidine of PmoB (His33) as well as His137 and His139 [Fig. 1A, M. capsulatus (Bath) numbering]. On the basis of extended x-ray absorption fine structure (EXAFS) data, this site was initially modeled as dicopper in some (10,11, 15), but not all (11–13), structures, with a later quantum refinement study supporting the monocopper assignment (16). LUntil now, it remained unclear whether the monocopper CuB site in the crystal structures is due to copper loss during the purification/crystallization process or whether CuB is actually a monocopper center, (iii) Last, a copper ion is found in the PmoC subunit coordinated by residues Asp156, His160, and His173(12).
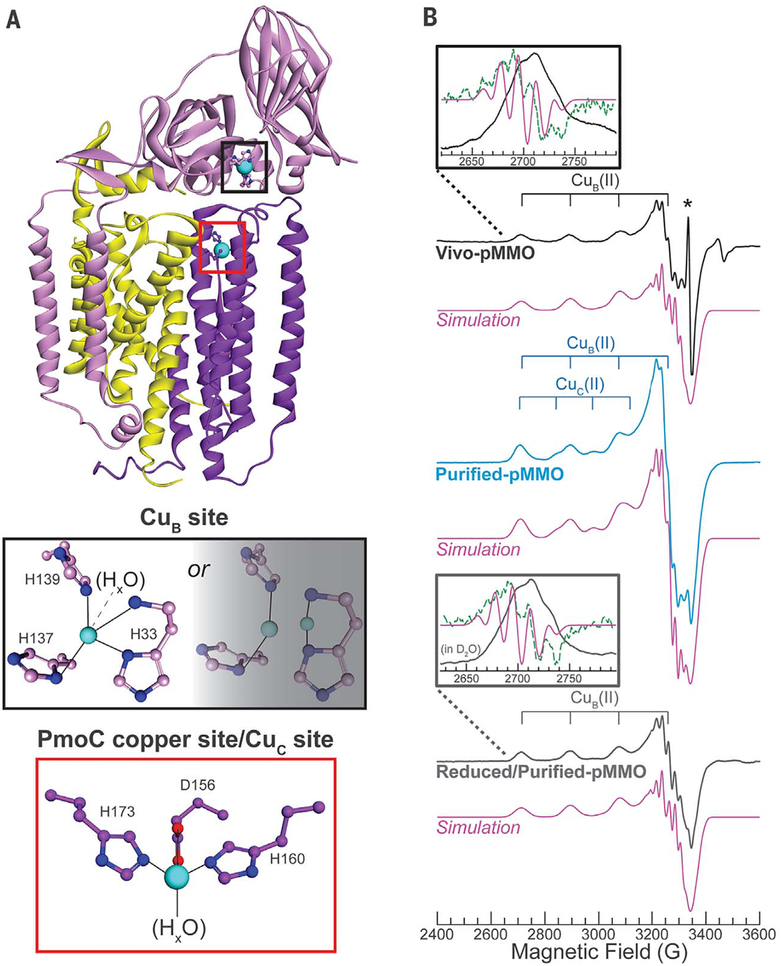
Fig. 1. Structure of one pMMO protomer as well as X-band continuous wave (CW) EPR of Vivo- [showing CuB(II)], Purified-[showing CuB(II) and Cuc(II)], and Reduced/Purified-pMMO [showing CuB(II)].
(A) (Top) Single protomer from the M. capsulatus (Bath) pMMO crystal structure (DOI: 10.2210/pdb3rgb/pdb) (11), showing PmoA (yellow), PmoB (pink), PmoC (purple), Cu (cyan), N (blue), and O (red) atoms. (Middle) The CuB site modeled as monocopper and dicopper. (Bottom) The PmoC metal site, which we have now determined to be the Cuc site, occupied with copper. (B) EPR spectra with simulations of the CuB(II) (Vivo- and Reduced/Purified-pMMO) and CuB(II) plus 0.32 equivalents Cuc(II) (Purified-pMMO) shown below each spectrum. (Insets) Lowest-field Cu hyperfine transition with computed second derivative (green dotted line) and second derivative of the simulation (pink solid line). Asterisk denotes an organic radical species in Vivo-pMMO. This radical is not present in Purified- or Reduced/Purified-pMMO. In the Reduced/Purified-pMMO (inset), the two lowest field 15N hyperfine lines are unresolved, likely because of a small amount of Cuc(II). Spectra and simulation parameters are listed in table S1, and collection conditions are provided in the supplementary materials. Rapid-passage Q-band absorption-display CW EPR spectra are shown in fig. S12. Unless otherwise noted, the concentrations of all EPR/ENDOR samples of Purified- or Reduced/Purified-pMMO were 300 to 500 μM. All pMMO spectra shown in the main text were measured on 63Cu, 15N-labeled pMMO samples.
The nuclearity, ligation, and location of the pMMO copper active site have been difficult to assign. The pMMO isolation and purification procedure has been suggested to result in loss or alteration of the essential metallocofactor, which is consistent with the substantially lower activity of pMMO after isolating the membranes from the organism (≲17% of that in vivo) (9,13). Variable metal content and enzymatic activity not having been demonstrated in crystals also call into question the physiological and catalytic relevance of the metallocofactors observed in crystal structures. Catalysis has been proposed to occur at three different types of multinuclear center, two of which have been dismissed: a tricopper site in PmoA (17–19), which is neither observed crystallographically nor by multiple investigators (including ourselves) with electron paramagnetic resonance (EPR) spectroscopy (20–25), and a diiron center at the PmoC metal-binding site (26), which is ruled out by the observation that copper, not iron, restores activity of metal-depleted pMMO (27). The third such proposal is that the active site is a dicopper CuB, located at the amino terminus of PmoB (27). This report addresses the Cu nuclearity in pMMO and shows that pMMO only contains two distinct monocopper sites. A detailed discussion and reconciliation of prior experiments (which were interpreted in terms of pMMO containing a dicopper center) with the present conclusion is provided in the supplementary materials.
To circumvent any complications arising from loss of copper cofactors through enzyme purification, we probed the pMMO Cu(II) sites in whole cells of M capsulatus (Bath) grown on 15N and 63Cu with EPR spectroscopy. Under copper-replete conditions, pMMO is highly expressed [~20% of total protein (26)]. Therefore, any Cu(II) sites within the protein should be present in such high quantity as to dominate the in vivo EPR spectrum. Indeed, prior EPR spectra of wholecell methanotroph samples exhibited a type 2 Cu(II) EPR signal with four N equatorial ligands that was attributed to pMMO-bound Cu(II) on the basis of the high Cu(II) concentration and similarity to the EPR spectrum of isolated methanotroph membranes (18,20,28). As a precursor to advanced spectroscopic characterization of the pMMO Cu(II), we first confirmed that we too observed this type 2 Cu(II) EPR signal, with four N equatorial ligands [as indicated by a five-line 15N hyperfine splitting of the low-field g1 Cu(II) hyperfine transition (fig. S2)], in the whole-cell (Vivo-pMMO) EPR spectrum of 15N, 63Cu-enriched M. capsulatus (Bath) (g = [2.242,2.068, 2.035]; 63Cu hyperfine splitting A1 = 570 MHz or 190 × 10−4 cm−1) (Fig. 1 and fig. S2). The ratio of g1/A1(cm−1) = 118 cm indicates a highly planar equatorial Cu coordination (29–31).
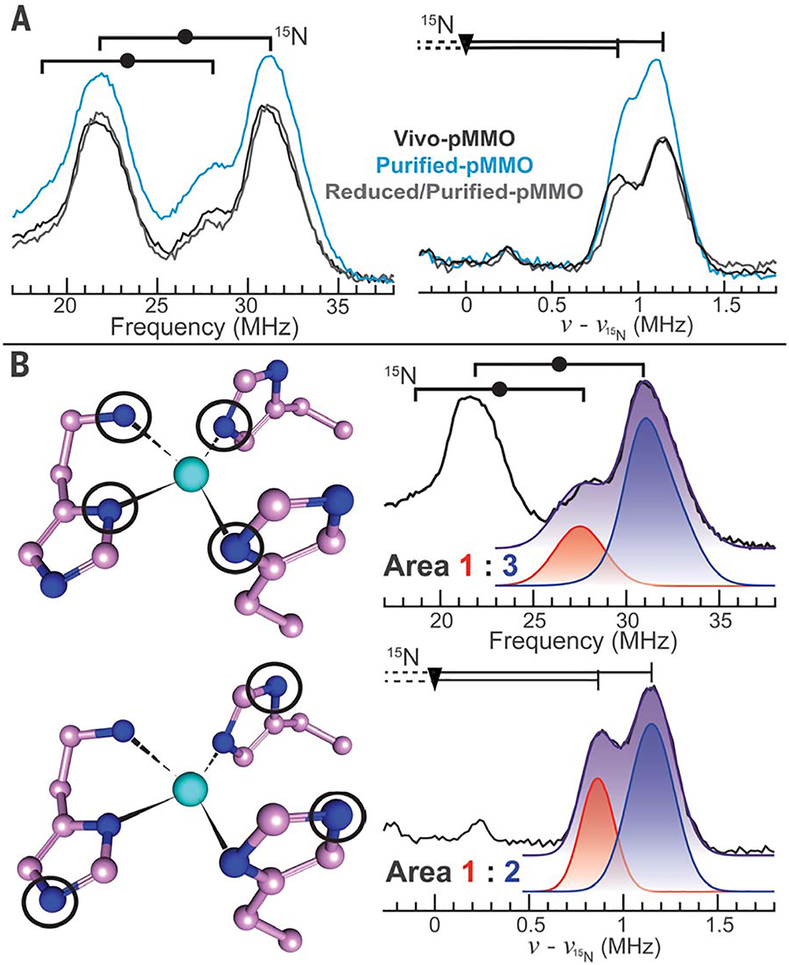
We characterized the nitrogenous ligands of this Cu(II) species with electron nuclear double resonance (ENDOR) spectroscopy. 15N Davies ENDOR spectra collected near g1 exhibited strongly coupled15N resonances corresponding to two similar directly coordinated Cu(II) ligands [|A1(15N)|~48,53 MHz] (Fig. 2A); Gaussian fitting and quantitation of the resonances indicated that the v+ peak with the larger coupling is three times more intense than the other v+ peak (Fig. 2B and tables S1 and S2). The near equivalence of these couplings combined with the quantitation is evidence for a Cu(II) site with four (determined by the 3:1 intensity ratio) equatorial 15N ligands bound in a square plane (32). 15N Doan/ReMims ENDOR spectra collected near also exhibit weakly coupled 15N resonances from the remote (noncoordinated) 15N of histidyl imidazoles bound to Cu(II) (|A1(15N)|~1.7, 2.3 MHz) (Fig. 2A) (33) in a 1:2 intensity ratio (Fig. 2B and tables S1 and S2). Thus, of the four N ligands defined by the 15N Davies ENDOR, three are histidyl imidazole side chains. Consistent with this assignment, 1H Davies ENDOR measurements showed nonexchangeable signals with couplings of A2 ~ 4.5 and 2.5 MHz, which is characteristic of the ring protons of Cu(II)-bound histidyl imidazole (fig. S3 and table S1) (34). Additional broad, exchangeable 1H signals with couplings |A1| ~ |A2| ~ 10 MHz are as expected for protons of −NH2 coordinated to Cu(II) (fig. S3 and table S1) (34).
Fig. 2. Characterization of Cub(II) and Cuc(II) 15N ligation with ENDOR.
(A) Q-band pulsed ENDOR measurements collected at g1for (left) strongly and (right) weakly coupled 15N nuclei, using Davies and Doan/ReMims ENDOR, respectively (Left) Goalpost widths indicate twice the 15N Larmor frequency v(15N), and the filled circles define half the hyperfine coupling magnitude (|A/2|). (Right) The triangle defines v(15N), and the distance from triangle to vertical line equals |A/2|. (B)(Left) Circled nitrogen atoms produce the observed ENDOR responses to the right of each CuB site. (Right) Overlay of Vivo-pMMO experimental spectra with individual (red and blue) and summed (purple) Gaussian functions used to quantitate 15N resonance peaks. Parameters are listed in tables S1 and S2, and collection conditions are provided in the supplementary materials. Although only the v+ peak is shown for the Vivo-pMMO weakly coupled 15N ENDOR response, both v−and v+ are shown in fig. S13.
To assign the location of this Cu(II) species, we examined the histidine residues in all pMMO crystal structures, looking for sites where three imidazoles and an −NH2 could simultaneously coordinate a Cu(II). The only location that can supply this spectroscopically defined ligand set is the CuB site (Fig. 1 and fig. S4), which provides three imidazole nitrogens from His33, His137, and His139 as well as the −NH2 of His33. The correspondence between this ligand assemblage and the EPR/ENDOR-defined ligation strongly supports assignment of CuB as a monocopper site.
To characterize any pMMO copper sites that might be maintained in vivo in the reduced Cu(I) state, we allowed them to air oxidize by solubilizing and purifying M. capsulatus (Bath) pMMO with size exclusion chromatography (Purified-pMMO). The CuB(II) signal persisted in Purified-pMMO, but another Cu(II) signal appeared, denoted Cuc (g = [2.30,2.07,2.05]; 63Cu hyperfine splitting A1 = 440 MHz or 147 × 10−4 cm−1) (Fig. 2), as previously observed in purified pMMO EPR samples (13,24). The larger Cuc(II) g1/A1 = 156 cm is characteristic of a distorted (flattened) tetrahedral geometry (35,36). In support of the conclusion that pMMO houses only monocopper centers, we have also found that an optical spectrum previously proposed to result from a dicopper center (37) is instead associated with a product of O2 or H2O2 oxidation of ascorbate in the presence of methanol and copper (fig. S5 and supplementary text).
Reduction of Purified-pMMO (Reduced/Purified pMMO) effectively eliminated the Cuc(II) EPR signal, leaving a CuB(II) signal virtually identical to that observed in vivo (Fig. 2), similar to previous reports of the reduction of pMMO localized in methanotroph membranes (18, 38), and with unchanged 15N ENDOR responses (Fig. 2), confirming that the CuB(II) site is unchanged during purification. The nonexchangeable signals from the imidazole ring protons and the exchangeable signals from protons attributed to −NH2 (fig. S3B) are also unchanged. We farther addressed the four-N ligation of CuB by EPR/ENDOR characterization of Reduced/Purified-pMMO incubated with H217O. The characteristic 17O ENDOR response of an equatorially coordinated HxO is absent, showing that CuB does not have such a ligand (fig. S6). The experiments did exhibit the 17O signal characteristic of an axial Hx17O on CuB, however, meaning that the geometry of CuB(II) is best described as square pyramidal.
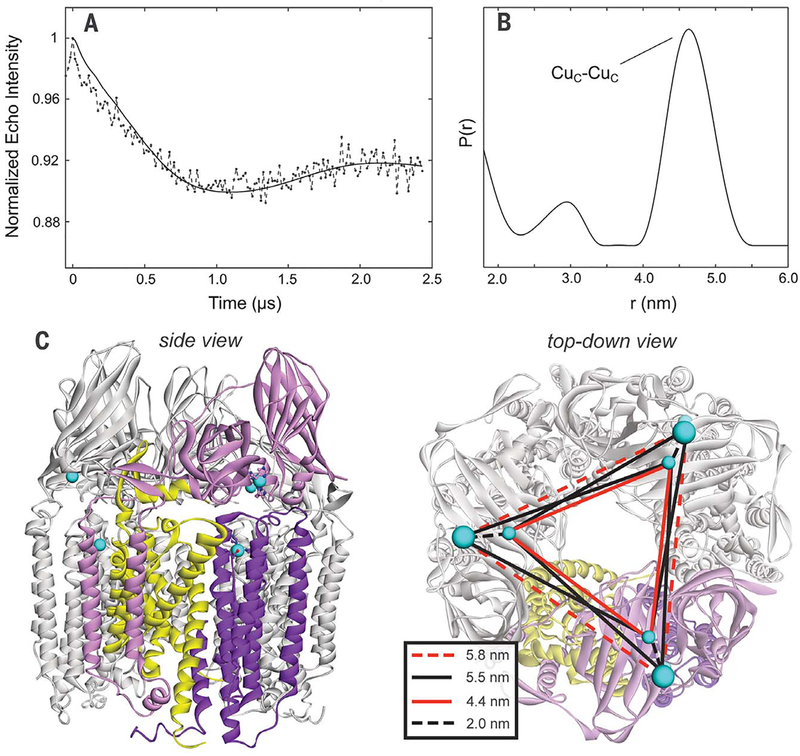
To determine the location of the Cuc(II) site, we measured Cu(II)-Cu(II) distances in Purified-pMMO with double electron-electron resonance (DEER) spectroscopy. The background-corrected DEER dipolar evolution for Purified-pMMO and corresponding Cu(II)-Cu(II) distance distribution obtained through Fourier transformation (39) are shown in Fig. 3. The transform of this 2.5-μs dipolar evolution can be considered robust for distances (d)of 2≲d≲5 nm. There are two peaks in the distance distribution: a major peak at a distance of 4.5 nm and a minor peak at 2.8 nm. The 2.8-nm peak is very weak and is extremely variable in intensity depending on the procedure used to analyze the time wave (fig. S7), whereas the 4.5-nm distance is robust, and indeed its counterpart can be clearly seen as the sinusoidal modulation of the time-evolution trace. Thus, we consider the shorter distance peak to be an artifact of the data processing.
Fig. 3. Purified-pMMO Cu(II)-Cu(II) four pulse DEER distance measurements.
(A) Normalized first-order homogeneous background decay-corrected dipolar evolution (solid line is a fitting of the dipolar evolution using DeerAnalysis2016). (B) Cu(II)–Cu(II) distance distributions calculated by using DeerAnalysis 2016. The measured Cu(II)–Cu(II) distance (4.5 nm) is consistent with the distances expected for Cuc(II) in the PmoC variable metal site and excludes the presence of Cu(II) in the bis-His site; if Cuc(II) was located in the bis-His site, a 3.2-nm Cu(II)—Cu(II) distance would be predicted (fig. S1). (C) Crystal structure of the M. capsulatus (Bath) pMMO trimer with same coloring as Fig. 1 (except two protomers are globally colored gray), and the various predicted Cu-Cu distances (inset).
We compared the 4.5-nm distance with the Cu-Cu distances in the M. capsulatus (Bath) pMMO crystal structure, which contains two metal-binding sites besides the CuB site: the bis-His site in PmoB (fig. S1) and the PmoC metal binding site (Fig. 1). If the Cuc(II) EPR signal corresponded to the bis-His site, a second robust Cu-Cu distance of ~3.2 nm (Cuc to Cuc) would be expected as well as the ~4.4-nm distance (in-terprotomer CuB to Cuc), contrary to observation. However, assignment of Cuc(II) to the PmoC site predicts a single Cu-Cu distance of −4.4 nm (Cuc to Cuc) (Fig. 3C), which is in agreement with the data. The intraprotomer ~2-nm CuB-to-Cuc distance distribution is not observed, presumably because it is too close to the minimum distance that can be resolved. Overall, the distance measurements indicate that Cue is not located in the bis-His site and instead is located in the PmoC metal binding site. There is no evidence of Cu(II) in the bis-His site in this sample or in any other structures [including one in which both histidines are present (13)], implying that this site may have been adventitious in the initial structure (10). This assignment is supported by 15N ENDOR measurements on Purified-pMMO (Fig. 3), which are consistent with the PmoC Cuc ligand assemblage depicted in Fig. 1. 1H and 17O ENDOR further indicate that Cuc contains a HxO ligand (figs. S3 and S6), as modeled in two pMMO crystal structures (11,12).
The characterization of these two monocopper sites in pMMO reopens the question of the identity of the catalytic site. The mononuclear CuB site exhibits saturated equatorial coordination, with strongly bound N-ligands, and thus is unlikely to undergo O2-binding/activation without alteration. However, addition of nitrite [a known inhibitor of methane oxidation (40,41)] perturbs the Cuc(II) EPR signal (fig. S8A), and ENDOR characterization of pMMO after addition of 15N-nitrite revealed a small 15N hyperfine coupling (A ~ 0.3 MHz), which is consistent with NO2− bound to Cu(II) through the oxygen(s) (fig. S8B) (42). This observation, combined with Verrucomicrobial pMMOs possessing none of the CuB-ligating amino acids (43), prompted us to reinvestigate the activity of the spmoB protein. This recombinantly expressed construct comprises the soluble portion of the M. capsulatus (Bath) PmoB subunit and is a model for native PmoB copper binding. It was reported to exhibit methane oxidation activities 1 to 10% that of pMMO (24,27), the only direct evidence identifying CuB as the active site.
To probe CuB reactivity in a more stable protein platform, we generated several fusion constructs of spmoB (fig. S9) that did not require refolding after expression [unlike spmoB, which expresses into inclusion bodies (27)]. One of these constructs assembled a CuB(II) site very similar to CuB(II) of pMMO by EPR (fig. S10A), unlike spmoB, which exhibits a different EPR spectrum (24). We tested the new constructs and the original spmoB for activity (figs. S10B and S11). These reaction mixtures produced 13C-methanol when assayed for 13C-methane oxidation. However, the amount of 13C-methanol produced was not affected by mutating the CuB site in the soluble spmoB construct; by altering the temperature, reaction time, protein concentration; or by using an unrelated copper enzyme incapable of oxidizing methane (fig. S11, C to E). The activity observed in these assays is instead attributed to the ability of duroquinol to reduce O2 and generate H2O2. H2O2 in turn can produce OH• through autolysis and through Fenton and Haber-Weiss chemistry (44–47), which then oxidizes methane. These experiments indicate that CuB in spmoB does not catalyze methane oxidation, thus eliminating the grounds for proposing CuB as the pMMO active site.
Instead, there is evidence that Cuc, located in the site illustrated in Fig. 1, may be the site of O2 binding and methane oxidation. Such a model would be consistent with the suggested presence of a displaceable solvent ligand on Cuc, as needed for O2-binding/activation, binding of the nitrite inhibitor to Cuc, and the absence of the CuB site in the Verrucomicrobial pMMOs. Last, mutation of any Cuc ligand in a pMMO homolog from Mycobacterium NBB4 (hydrocarbon monooxygenase) resulted in complete loss of activity (48). Cuc is thus inferred to be the active site of hydrocarbon substrate binding and oxidation, but CuB nonetheless is important. Replacement of a CuB ligand in the homolog diminished overall activity significantly (by 80%), but the variant’s affinity for alkane substrate was within error of wild type for two of the three alkanes assayed (48), indicating that CuB does play a functional role, even though it is not the site of hydrocarbon substrate binding and oxidation.
Supplementary Material
ACKNOWLEDGMENTS
We thank G. E. Cutsail III, M. A. Culpepper, and H. W. Pinkett for helpful discussions as well as an anonymous reviewer for the expert analysis of the Gaussian fitting and providing above and beyond effort.
Funding: This work was supported by NIH grants GM118035 (A.C.R.), GM111097 (B.M.H.), and 5T32GM008382 (M.O.R.) and NSF grant 1534743 (S.L.M., B.D.O., and A.C.R.). F.M. was supported by a Royal Society Wolfson Research Merit Award.
Footnotes
Competing interests: A patent related to this work has been issued: A. C. Rosenzweig, T. J. Lawton, A. Nisthal, J. S. Kostecki, H. K. Privett, F. Lee, B. Olafson, A. D. Dousis, “Engineered recombinant enzymes for methane oxidation” U.S. patent US9896700B2. B.D.O. is a cofounder and the CEO of Protabit, a for-profit company that develops and markets software for protein engineering and computational protein design. S.L.M. is a cofounder of Protabit.
Data and materials availability: All data are available in the manuscript or the supplementary materials.
SUPPLEMENTARY MATERIALS
science.sciencemag.org/content/364/6440/566/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S16
Tables S1 to S3
References (49–68)
REFERENCES AND NOTES
- 1.Ross MO, Rosenzweig AC, J. Biol. Inorg. Chem 22, 307–319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandt AR et al. , Science 343, 733–735 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Environmental Protection Agency (EPA), Inventory of U.S. greenhouse gas emissions and sinks: 1990-2015 (EPA, 2017). [Google Scholar]
- 4.Fei Q et al. , Biotechnol. Adv 32, 596–614 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Horn R, Schlögl R, Catal. Lett 145, 23–39 (2015). [Google Scholar]
- 6.Snyder BER, Bols ML, Schoonheydt RA, Sels BF, Solomon EI, Chem. Rev 118, 2718–2768 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Khan MS, Park JH, Chaniago YD, Lee M, Energy Procedia 61, 599–602 (2014). [Google Scholar]
- 8.Gao J et al. , Science 348, 686–690 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Lawton TJ, Rosenzweig AC, J. Am. Chem. Soc 138, 9327–9340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lieberman RL, Rosenzweig AC, Nature 434, 177–182 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Smith SM et al. , Biochemistry 50, 10231–10240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sirajuddin S et al. , J. Biol. Chem 289, 21782–21794 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ro SY et al. , J. Biol. Chem 293, 10457–10465 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balasubramanian R, Rosenzweig AC, Acc. Chem. Res 40, 573–580 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Hakemian AS et al. , Biochemistry 47, 6793–6801 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao L, Caldararu O, Rosenzweig AC, Ryde U, Angew. Chem. Int. Ed 57, 162–166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang VCC et al. , Chem. Rev 117, 8574–8621 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Nguyen HH et al. , J. Biol. Chem 269, 14995–15005 (1994). [PubMed] [Google Scholar]
- 19.Chan SI et al. , Angew. Chem. Int. Ed 46, 1992–1994 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Yuan H, Collins MLP, Antholine WE, J. Am. Chem. Soc 119, 5073–5074 (1997). [Google Scholar]
- 21.Choi DW et al. , Microbiology 151, 3417–3426 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Basu P, Katterle B, Andersson KK, Dalton H, Biochem. J 369, 417–427 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lieberman RL et al. , Proc. Natl. Acad. Sei. U.S.A. 100, 3820–3825 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Culpepper MA, Cutsail GE III, Gunderson WA, Hoffman BM, Rosenzweig AC, J. Am. Chem. Soc 136, 11767–11775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zahn JA, DiSpirito AA, J. Bacteriol 178, 1018–1029 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinho M et al. , J. Am. Chem. Soc 129, 15783–15785 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balasubramanian R et al. , Nature 465, 115–119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemos SS, Perille Collins ML, Eaton SS, Eaton GR, Antholine WE, Biophys. J 79, 1085–1094 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakaguchi U, Addison AW, J. Chem. Soc 600–608 (1979). [Google Scholar]
- 30.Pogni R, Baratto MC, Diaz A, Basosi R, J. Inorg. Biochem 79, 333–337 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Patel RN et al. , Inorg. Chim. Acta 362, 4891–4898 (2009). [Google Scholar]
- 32.Iwaizumi M, Kudo T, Kita S, Inorg. Chem 25, 1546–1550 (1986). [Google Scholar]
- 33.Dicus MM et al. , J. Am. Chem. Soc 132, 2037–2049 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manikandan P, Epel B, Goldfarb D, Inorg. Chem 40, 781–787 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Yokoi H, Bull. Chem. Soc. Jpn 47, 3037–3040 (1974). [Google Scholar]
- 36.Cheeseman TP, Hall D, Waters TN, J. Chem. Soc. A 1966, 685–693 (1966). [Google Scholar]
- 37.Culpepper MA, Cutsail III GE, Hoffman BM, Rosenzweig AC, J. Am. Chem. Soc 134, 7640–7643 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan H, Collins ML, Antholine WE, J. Inorg. Biochem 72, 179–185 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Jeschke G et al. , Appl. Magn. Reson 30, 473–498 (2006). [Google Scholar]
- 40.Nyerges G, Stein LY, FEMS Microbiol. Lett 297, 131–136 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Stein LY, Arp DJ, Appl. Environ. Microbiol 64, 4098–4102 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fittipaldi M et al. , Biochemistry 44, 15193–15202 (2005). [DOI] [PubMed] [Google Scholar]
- 43.Op den Camp HJ et al. , Environ. Microbiol. Rep 1, 293–306 (2009). [DOI] [PubMed] [Google Scholar]
- 44.White CC, Chain RK, Malkin R, Biochim. Biophys. Acta 502, 127–137 (1978). [DOI] [PubMed] [Google Scholar]
- 45.Taylor CE, Prepr. Pap.-Am. Chem. Soc. Div. Fuel Chem. 48, 876 (2003). [Google Scholar]
- 46.Cadenas E, Boveris A, Ragan CI, Stoppani AOM, Arch. Biochem. Biophys 180, 248–257 (1977). [DOI] [PubMed] [Google Scholar]
- 47.Ikai H et al. , Antimicrob. Agents Chemother. 54, 5086–5091 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liew EF, Tong D, Coleman NV, Holmes AJ, Microbiology 160, 1267–1277 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.