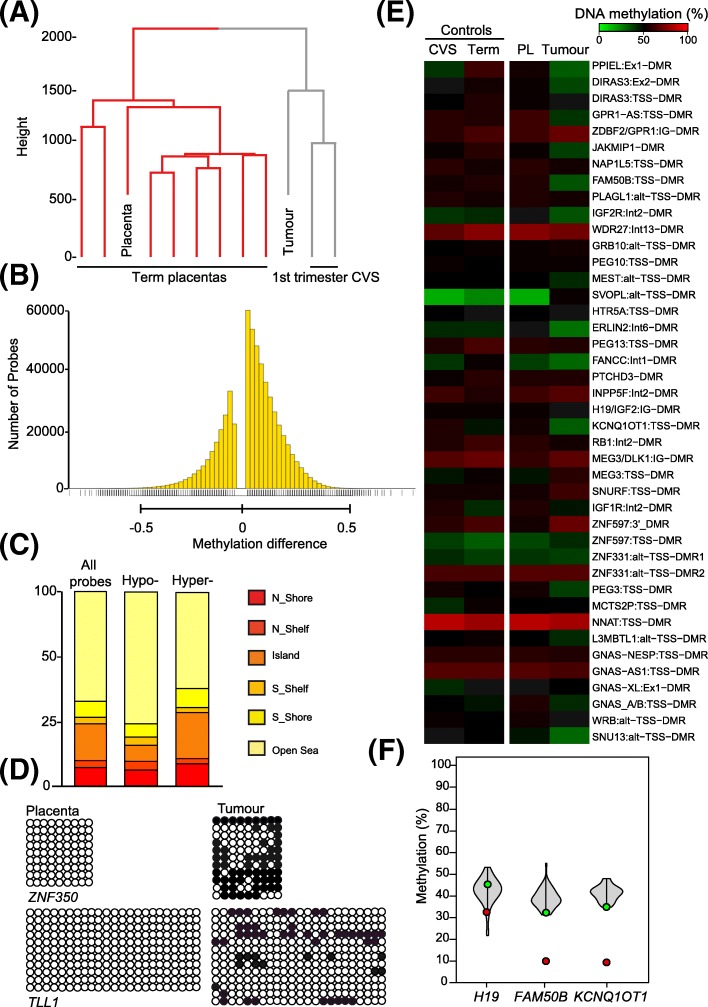
Fig. 2.
Characterization of DNA methylation in the placenta and tumour using the Illumina EPIC methylation array. a Hierarchical clustering of global methylation for placenta-tumour paired samples with 8 term placenta and 2 first trimester CVS samples. b Bar graph of the distribution of the hypo- (left side) and hypermethylated (right side) probes with a difference greater that −/+ 2.5% (0.025ß) when comparing the tumour and paired placenta. c Classification of probes with differential methylation according to genomic location. The bar chart illustrates probe enrichment classified by Illumina Infinium annotation. d Bisulphite confirmation of methylation difference between tumour and paired placenta samples. Each circle represents a single CpG dinucleotide on a DNA strand. (•) Methylated cytosine, (o) unmethylated cytosine. Each row corresponds to an individual cloned sequence. e Heatmap of the ubiquitous imprinted DMR probes in control first trimester CVS and term placenta biopsies as well as the tumour and paired placenta samples. The values represent the average of all probes mapping to each region. f Methylation levels of ubiquitous imprinted DMRs in the placenta (green dot), tumour (red dot) and controls (violin plots n = 16) quantified by pyrosequencing