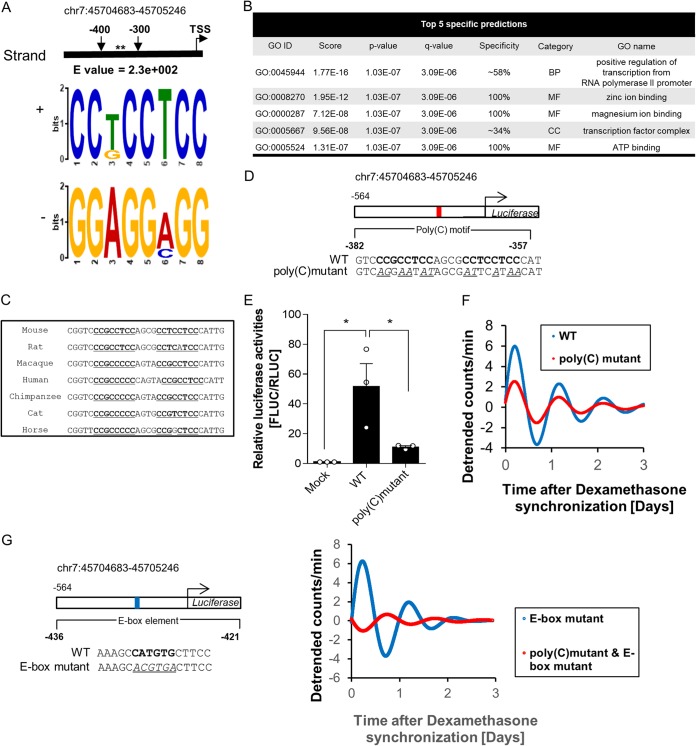
FIG 2.
The poly(C) motif is critical for Dbp promoter activity. (A) The poly(C) motif (or CCT motif) was scanned in the region between bp −300 and −400 from the TSS using the MEME suite. The asterisks mark the position of the poly(C) motif. (B) One of the functions of the poly(C) motif predicted by GOMo was transcription. The poly(C) motif was the only motif that was predicted to function in transcription. MF, molecular function; BP, biological process; CC, cellular component. (C) Alignment of genomic regions containing a pair of poly(C) motifs showed high conservation among eutherian mammals, which suggests that a common transcription-regulatory mechanism is involved in Dbp promoter activity. Poly(C) motifs are boldface and underlined. (D) Diagram of the luciferase reporter system containing a WT promoter and a poly(C) mutant. The poly(C) motif is in boldface; mutated bases are underlined and italic. (E) Promoter activity 24 h after dexamethasone synchronization of the poly(C) mutant showed a significant decrease compared to the WT, which suggests that the poly(C) motif is crucial for promoter activity (n = 3; *, P < 0.05 by two-way ANOVA followed by Tukey's multiple-comparison test). The bars indicate means and SEM. (F) Representative detrended bioluminescence recordings of WT:LUC and poly(C) mutant:LUC measured with a luciferase reporter-transfected NIH 3T3 cell line after dexamethasone synchronization, which showed a decrease in the amplitude level of the poly(C) mutant. (G) Representative detrended bioluminescence recordings of an E-box mutant:LUC and an E-box and poly(C) mutant:LUC measured with a luciferase reporter-transfected NIH 3T3 cell line after dexamethasone synchronization. The data showed a dramatic decrease in the amplitude level of the E-box and poly(C) mutant promoter.