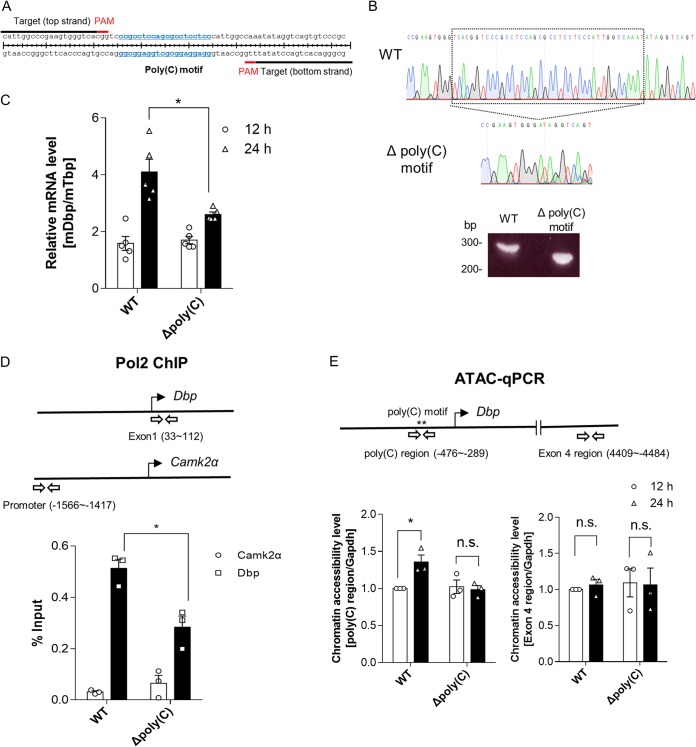
FIG 3.
The poly(C) motif is important for RNA polymerase 2 recruitment because it induces greater chromatin accessibility. (A) Schematic depiction of a pair of sgRNAs designed to delete a pair of endogenous poly(C) motif regions within the Dbp proximal promoter region. (B) A cell line with the poly(C) motif region deleted made with the CRISPR-Cas9 system was confirmed by genotyping and sequencing. The boxed region is between bp −397 and −337 from the TSS. (C) The endogenous Dbp mRNA was quantified, and the cell line with the poly(C) motif region deleted showed decreases in the mRNA expression level at 24 h after dexamethasone synchronization (n = 5; *, P < 0.05 by two-way ANOVA followed by Tukey's multiple-comparison test). (D) Pol2 ChIP was conducted to measure the recruitment level of Pol2 to Dbp regions at 24 h after dexamethasone synchronization (n = 3; *, P < 0.05 by two-way ANOVA followed by Tukey's multiple-comparison test). (E) ATAC-qPCR data indicated that chromatin accessibility of WT cells increased 24 h after dexamethasone synchronization; however, cells with the poly(C) motif region deleted did not show an increase in chromatin accessibility. Furthermore, the Dbp exon 4 region was not significantly changed. The Gapdh promoter region was used for normalization, which showed no oscillatory patterns of mRNA expression, in contrast to Dbp mRNA (n = 3; n.s., not significant; *, P < 0.05 by two-way ANOVA followed by Tukey's multiple-comparison test). The bars indicate means and SEM.