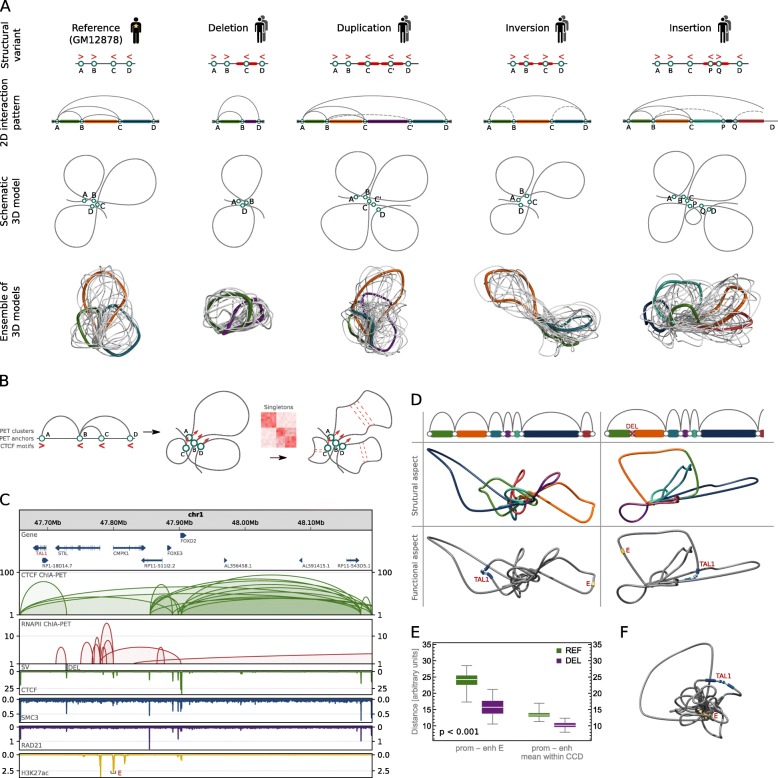
Fig. 3.
Computational algorithm for modeling topological alterations caused by SVs. a Predicted impact of particular SV types on looping structure of the genome. Simplified chromatin looping patterns and 3D models are presented for the reference and its SV-altered versions. b Scheme presenting the chromatin modeling method at the level of loops. The method uses PET clusters, singletons, and orientations of CTCF binding motifs to accurately model the genome looping structures. c Browser view of a topological domain containing TAL1 gene and a deletion causing its activation. The deletion removes CTCF insulating the TAL1 promoter from enhancer E. CTCF and RNAPII ChIA-PET interactions are shown along with ChIP-seq tracks for CTCF, cohesin subunits (SMC3 and RAD21), and H3K27ac which marks the enhancer E. d Models presenting 3D structure of the TAL1 locus without the deletion (left column) and with the deletion (right column). Schematic drawings of loops shown in c (first row); 3D models with loops colored as on schematic drawings (second row); 3D models with TAL1 and enhancer E marked (third row). e Distance in 3D Euclidian space between the TAL1 promoter and enhancer E and mean distance between the promoter and enhancers located in the same CCD. In green, distribution of distances calculated in 3D models of the reference structure (REF), in purple—in models with the deletion introduced (DEL). For each case, 100 models were generated. The differences between REF and DEL groups are statistically significant (p values much less than 0.001), see Fig. 2c for box plot description. f 3D model of the TAL1 locus including RNAPII-mediated chromatin interactions