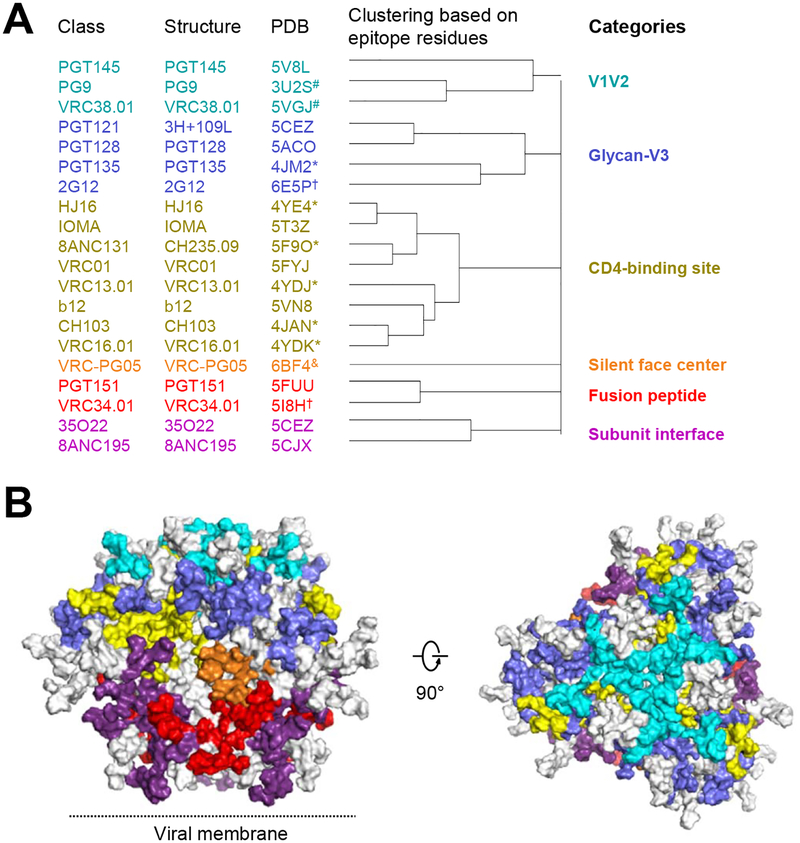
Figure 1. 20 classes of broadly neutralizing antibodies recognize the prefusion-closed Env trimer and segregate into 6 categories based on the Env residues with which they interact.
(A) All HIV-1 Env-antibody complexes structures in the PDB (Table 1) were assigned to classes (leftmost column, listed by the name of first reported antibody of the class) based on similarities in B cell ontogeny and mode of recognition, with a representative structure and PDB for the class (2nd and 3rd columns from left), which were chosen based on resolution and degree to which Env in the structure resembled prefusion-closed trimer; “*” indicate structures determined in deglycosylated gp120-core context; “&” indicates structures determined in partially glycosylated gp120-core context; “#” indicates structures determined with V1/V2 scaffold; “†”indicates a structure with high resolution peptide- or glycan-antibody complex but lower resolution Env trimer-antibody complex structure; “@” indicates glycan N295, N332, and N392 used for 2G12 epitope (see methods). (B) Prefusion-closed Env trimer with molecular surface colored by categories defined in (A). Epitope residues shared by antibodies in separate classes were colored according to surface area and requirements for antibody binding. For example, glycan N276 has interactions with antibody 8ANC195 of the subunit interface category and with multiple antibodies of the CD4-binding site category; however, because glycan N276 is required for 8ANC195, and generally only accommodated by antibodies that target the CD4-binding site, we colored glycan N276 to be part of the subunit interface category. Left image is shown with viral membrane at bottom; right image is rotated 90° to look down on the trimer apex. See also Figures S1 and S5, Tables S1 and S3, and Dataset S1.