Figure 10.
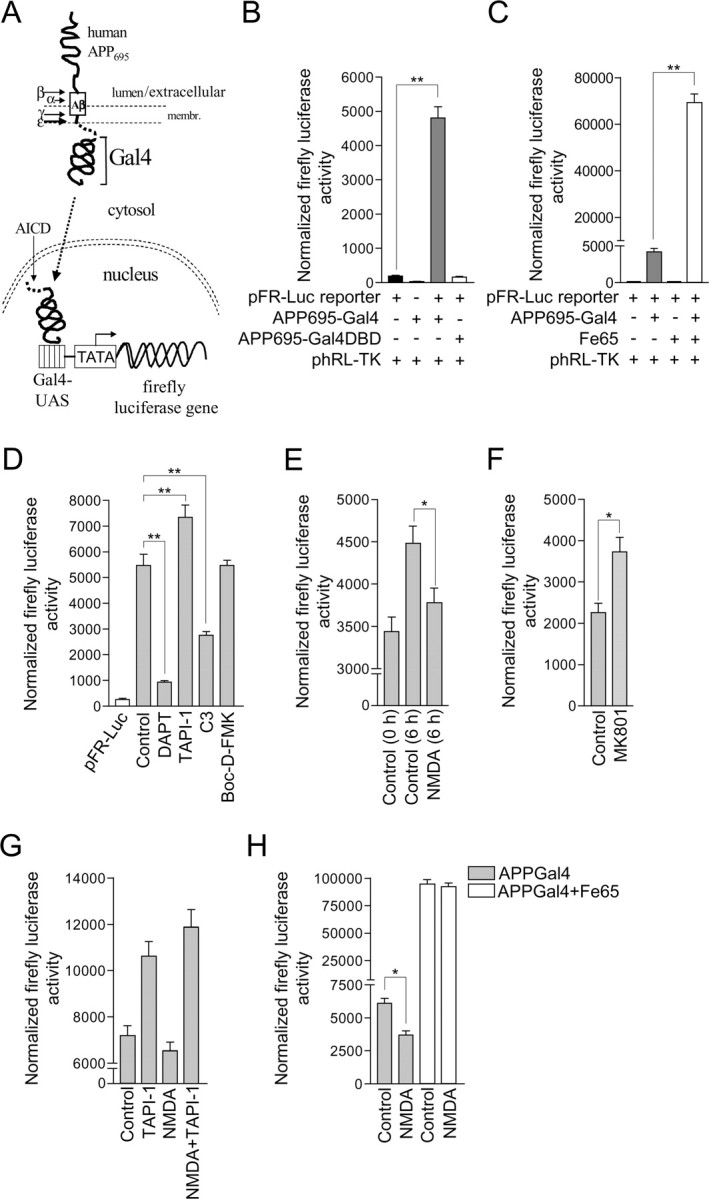
NMDA receptor stimulation inhibits AICD-Gal4-driven luciferase reporter gene activity in primary cultured cortical neurons. A, Schematic representation of a cell-based reporter gene assay for γ-secretase-mediated cleavage of APP in primary cultured cortical neurons. The assay uses a reporter APP protein consisting of human APP695 fused at its C terminus to the yeast transcription factor Gal4. After proteolytic processing of exogenously expressed APP695-Gal4 by α-secretase and/or β-secretase, cleavage by γ-secretase at the ε-site, produces an AICD-Gal4 fragment that can translocate to the nucleus in which Gal4 induces transcription of a transfected Gal4-dependent firefly luciferase reporter gene. Firefly luciferase expression is quantified by performing a Dual-Glo luciferase activity assay as detailed in Materials and Methods. B, Primary cultured cortical neurons at 8 DIV were transfected with pFR-Luc firefly luciferase reporter gene plasmid alone and in combination with plasmids encoding for APP695-Gal4 or APP695-Gal4DBD. All cells were cotransfected with phRL-TK plasmid that constitutively expresses moderate levels of Renilla luciferase. Dual-Glo luciferase activity assays were performed 24 h after transfection for quantification of firefly and Renilla luciferase expression. Firefly luciferase reporter activity was normalized using the constitutive Renilla luciferase activity. Each column is the mean ± SEM of 12 separate transfections (n = 12; **p < 0.01, pFR-Luc reporter vs pFR-Luc reporter+APP695-Gal4, one-way ANOVA with Bonferroni post test). C, Primary cultured cortical neurons at 8 DIV were transfected with pFR-Luc firefly luciferase reporter gene plasmid alone and in combination with plasmids encoding for APP695-Gal4 or Fe65. All cells were cotransfected with phRL-TK plasmid that constitutively expresses moderate levels of Renilla luciferase. Dual-Glo luciferase activity assays were performed 24 h after transfection for quantification of firefly and Renilla luciferase expression. Firefly luciferase reporter activity was normalized using the constitutive Renilla luciferase activity. Each column is the mean ± SEM of 12 separate transfections (n = 12; **p < 0.01, pFR-Luc reporter+APP695-Gal4 vs pFR-Luc reporter+APP695-Gal4+Fe65, one-way ANOVA with Bonferroni post test). D, Primary cultured cortical neurons at 8 DIV were treated with vehicle (control), 10 μm DAPT, 50 μm TAPI-1, 10 μm C3, or 20 μm Boc-d-FMK for 30 min before cotransfection with pFR-Luc firefly luciferase reporter gene, APP695-Gal4 and phRL-TK plasmids. Inhibitors were present throughout the course of the experiment. Dual-Glo luciferase activity assays were performed 24 h after transfection for quantification of firefly and Renilla luciferase expression. Firefly luciferase reporter activity was normalized using the constitutive Renilla luciferase activity. Inhibitor treatments did not alter the low level basal luciferase reporter activity arising from transfection of neurons with pFR-Luc reporter gene plasmid alone (data not shown). Each column is the mean ± SEM of 12 separate transfections (n = 12; **p < 0.01, control vs DAPT, control vs TAPI-1 control vs C3, one-way ANOVA with Bonferroni post test). E, Primary cultured cortical neurons at 8 DIV were cotransfected with pFR-Luc firefly luciferase reporter gene, APP695-Gal4, and phRL-TK plasmids. To determine whether NMDA receptor activity altered firefly luciferase reporter gene transcription over a 6 h time period, firefly and Renilla luciferase activity measurements were made in a subset of neurons 24 h after transfection (control 0 h), and at the same time, neurons were treated with vehicle (control) or 50 μm NMDA, and left for an additional 6 h followed by firefly and Renilla luciferase activity measurements. Firefly luciferase reporter activity was normalized using the constitutive Renilla luciferase activity. This experimental protocol is made possible because over the time course of the experiment, luciferase expression is cumulative and stable. Each column is the mean ± SEM of 56 separate transfections (n = 56; *p < 0.05, control vs NMDA, unpaired two-tailed Student's t test). F, Primary cultured cortical neurons at 8 DIV were cotransfected with pFR-Luc firefly luciferase reporter gene, APP695-Gal4 and phRL-TK plasmids. Twenty-four hours after transfection, neurons were treated with vehicle (control) or 2 μm MK801 for 6 h, followed by quantification of firefly and Renilla luciferase expression by performing Dual-Glo luciferase activity assays. Firefly luciferase reporter activity was normalized using the constitutive Renilla luciferase activity. Each column is the mean ± SEM of 56 separate transfections (n = 56; *p < 0.05, control vs MK801, unpaired two-tailed Student's t test). G, Primary cultured cortical neurons at 8 DIV were cotransfected with pFR-Luc firefly luciferase reporter gene, APP695-Gal4 and phRL-TK plasmids. Twenty-four hours after transfection, neurons were treated with vehicle (control), 50 μm TAPI-1, 50 μm NMDA, or 50 μm NMDA in the presence of 50 μm TAPI-1 (NMDA+TAPI-1) for 6 h, followed by quantification of firefly and Renilla luciferase expression by performing Dual-Glo luciferase activity assays. Firefly luciferase reporter activity was normalized using the constitutive Renilla luciferase activity. Each column is the mean ± SEM of 36 separate transfections. H, Primary cultured cortical neurons at 8 DIV were transfected with pFR-Luc firefly luciferase reporter gene plasmid in combination with APP695-Gal4 plasmid, or APP695-Gal4 and Fe65 plasmids. All cells were cotransfected with phRL-TK plasmid. Twenty-four hours after transfection, neurons were treated with vehicle (control) or 50 μm NMDA for 6 h, followed by quantification of firefly and Renilla luciferase expression by performing Dual-Glo luciferase activity assays. Firefly luciferase reporter activity was normalized using the constitutive Renilla luciferase activity. Each column is the mean ± SEM of 12 (APPGal4) or 24 (APPGal4+Fe65) separate transfections (*p < 0.05, APPGal4, control vs NMDA, unpaired two-tailed Student's t test).
