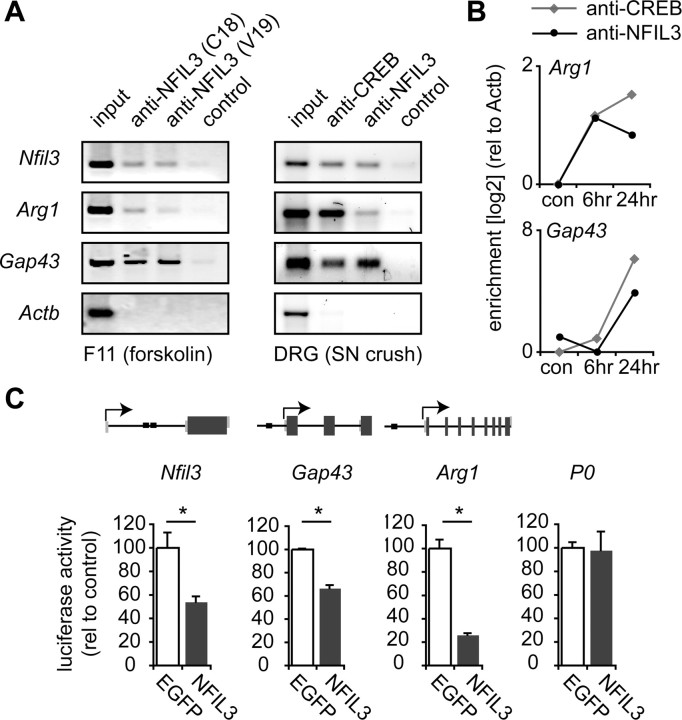
Figure 6.
NFIL3 binds to and represses transcription of regeneration-associated genes. A, ChIP assay showing NFIL3 binding to promoter regions of Nfil3, Arg1 and Gap43 in F11 cells overexpressing NFIL3 (left panel) and in DRG neurons 2 d after sciatic nerve crush in vivo (right panel). Two different NFIL3 antibodies were used that both precipitated all three promoters. No immunoprecipitation was observed in the absence of antibodies (control) or for a negative control gene (Actb). B, Quantitative PCR analysis of ChIP enrichment showing phospho-CREB and NFIL3 occupancy at the Arg1 and Gap43 promoters at 6–24 h after sciatic nerve crush. No binding was observed in unlesioned control DRGs. C, Schematic diagrams of the Nfil3, Gap43 and Arg1 genes indicating predicted EBPRE sites (black boxes). Indicated are coding exon regions (dark gray boxes), untranslated exon regions (light gray boxes), introns (black lines) and transcription start sites (arrows). Genomic fragments of the Nfil3, Gap43 and Arg1 genes containing predicted EBPRE sites were cloned into the pGL2-B-luciferase plasmid. Luciferase assays show that these constructs are transcriptionally active in HEK293 cells (white bars), and that transcriptional activity is repressed when NFIL3 is cotransfected (gray bars). Activity of the P0 promotor is not repressed by NFIL3. Bars represent means ± SEM; n = 3 for each condition; *p < 0.01.