Abstract
The oscillator mouse (Glra1spd-ot) carries a 9 bp microdeletion plus a 2 bp microinsertion in the glycine receptor α1 subunit gene, resulting in the absence of functional α1 polypeptides from the CNS and lethality 3 weeks after birth. Depending on differential use of two splice acceptor sites in exon 9 of the Glra1 gene, the mutant allele encodes either a truncated α1 subunit (spdot-trc) or a polypeptide with a C-terminal missense sequence (spdot-elg). During recombinant expression, both splice variants fail to form ion channels. In complementation studies, a tail construct, encoding the deleted C-terminal sequence, was coexpressed with both mutants. Coexpression with spdot-trc produced glycine-gated ion channels. Rescue efficiency was increased by inclusion of the wild-type motif RRKRRH. In cultured spinal cord neurons from oscillator homozygotes, viral infection with recombinant C-terminal tail constructs resulted in appearance of endogenous α1 antigen. The functional rescue of α1 mutants by the C-terminal tail polypeptides argues for a modular subunit architecture of members of the Cys-loop receptor family.
Keywords: chloride channel, domain, glycine receptor, TM3–TM4 loop, rescue, inhibition
Introduction
Glycine receptors (GlyRs) are a family of ligand-gated chloride channels that mediate neuronal inhibition throughout the CNS. The isoform GlyRA, which prevails in the spinal cord of adult rodents, is thought to be a heteropentameric complex of two α and three β subunits (Grudzinska et al., 2005). Although GlyRα2 homopentamers were described to constitute the isoform GlyRN of neonatal spinal cord (Becker et al., 1988; Hoch et al., 1989), α2 has also been observed in adult tissues, e.g., cerebellum and retina (Young and Cepko, 2004). Additional GlyR heterogeneity arises from gene variants of the ligand binding subunit (α3/α4) (Harvey et al., 2004; Heinze et al., 2007). The GlyR belongs to the superfamily of Cys-loop receptors, which also includes nicotinic acetylcholine, 5-HT3, and GABAA/C receptors (Lynch, 2004; Betz and Laube, 2006). Members of this superfamily share an Ig-like fold of the N terminus and four transmembrane domains (TMs), with TM2 lining the ion pore. Within the intracellular TM3–TM4 loop, a basic motif was identified mediating accumulation of receptors at the cell surface (Sadtler et al., 2003; Yevenes et al., 2006).
Hereditary hyperekplexia, or startle disease, is a human neurological disorder characterized by excessive startle responses and muscle stiffness in infancy. The disorder is associated with mutations of the GLRA1 gene (Rees et al., 1994; Becker et al., 2006, 2008), the β subunit gene GLRB (Rees et al., 2002), and the gene encoding the glycine transporter GlyT2 (Eulenburg et al., 2006; Rees et al., 2006). The mouse mutants oscillator (Glra1spd-ot) (Buckwalter et al., 1994; Kling et al., 1997) and spasmodic (Glra1spd) (Ryan et al., 1994; Saul et al., 1994) carry orthologous mutations in the Glra1 gene, whereas the spastic (Glrbspa) mouse carries a β subunit mutation (Becker et al., 1992; Kingsmore et al., 1994; Mülhardt et al., 1994). Two weeks after birth, oscillator mice develop a progressively worsening tremor and muscle spasms, resulting in death at the age of 3 weeks (Buckwalter et al., 1994; Büsselberg et al., 2001). The oscillator allele Glra1spd-ot exhibits a microdeletion of 9 bp plus a microinsertion of 2 bp. In the CNS of homozygous mutants, a complete loss of functional α1 protein is observed (Kling et al., 1997), characterizing oscillator as a functional null mutation of the Glra1 gene.
Depending on differential use of two splice acceptor sites in exon 9 of Glra1 (Malosio et al., 1991; Buckwalter et al., 1994), two α1 polypeptide mutants are encoded, both of which lack TM4. Inclusion of the alternative insert produces the truncated α1 variant, spdot-trc, attributable to a premature STOP codon. The long splice variant, spdot-elg, results from an exclusion of the alternative 24 nt and encodes 150 missense residues at the C terminus. To test our hypothesis that segments of the GlyRα1 subunit behave as self-organizing domains, a complementation of ion channel function was attempted by recombinant coexpression of oscillator α1 mutants with a C-terminal rescue construct, without being covalently attached to each other. Coexpression of the mutant subunit spdot-trc, or related constructs, with C-terminal segments of the wild-type (wt) α1 subunit, reconstituted glycine-gated chloride channels. Rescue efficiency was enhanced by a basic motif within the TM3–TM4 loop of the wild type. Although native neurons cultured from spinal cord of homozygous mutants lack the α1 subunit, expression of the tail construct resulted in the appearance of endogenous spdot antigen. The formation of functional GlyR channels from independent α1 subunit segments suggests a modular architecture of Cys-loop receptor subunits.
Materials and Methods
Chemicals.
Unless stated otherwise, all chemicals were from Sigma, Roth, or Invitrogen.
RNA isolation and reverse transcription-PCR.
RNA extraction from tissue and transfected cells was done as recommended by the manufacturer (PEQLAB) using peqGOLD RNAPure. One microgram of RNA was used for reverse transcription (RT)-PCR. Following a typical RT-PCR protocol, we used Moloney murine leukemia virus RT (H−) (50–100 units) provided with 5× reaction buffer and dATP, dCTP, dGTP, dTTP (10 mm each), and random hexamers (50–200 ng) (Promega). Two microliters of cDNA were used for every PCR reaction (95°C for 5 min; 25 cycles at 95°C for 30 s, 55°C for 30 s, 72°C for 30 s; 72°C for 10 min) for amplification of the housekeeping gene β-actin, the glycine receptor α1, α2, the C-terminal tail construct, and green fluorescent protein (GFP). The following primers were used for β-actin: β-actin sense (S), 5′-TGAGACCTTCAACACCCCAG-3′ and β-act antisense (AS), 5′-CATCTGCTGGAAGGTGGACA-3′; for GlyRα1: mma1-S, 5′-CCTTCTGGATCAACATGGATGCTG-3′ and mma1-AS, 5′-CGCCTCTTCCTCCTAAATCGAAGCAGT-3′; for GlyRα2: mma2-S, 5′-GGGTACACCATGAATGACCTG-3′ and mma2-AS, 5′-TAGCATCTGCATCTTTGGGGGGT-3′; for the C-terminal tail construct myc–α1–iD–TM4, with “iD” indicating the intracellular domain: tail-S, 5′-CTAGCCGCCGCCACCATGTGGACCCCGCGA-3′ and tail-AS, 5′-GGGGTACCTCACTTGTTGTGGACAT-3′; and for discrimination between the expanded (spdot-elg) and truncated (spdot-trc) α1 transcript: mma1-S, 5′-CTGCTTCGATTTAGGAGGAAGAGGCGA-3′ and mma1-AS, 5′-CAGAGATGCCATCCTTGG-3′. PCR fragments for spdot-elg and spdot-trc were cloned into the plasmid pRK7 and sequenced using an ABI Sequencer (Applied Biosystems) (Fig. 1C,D).
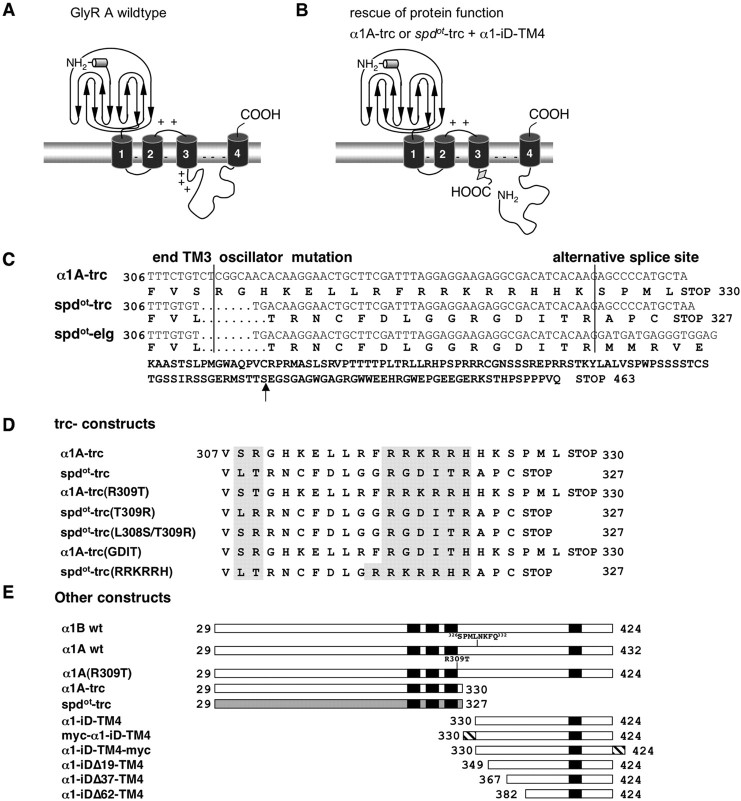
Figure 1.
GlyR constructs used. A, Topology model of GlyRs. Each receptor subunit is composed of four TMs, an extracellular N terminus, and an extracellular C terminus. TMs are connected by intracellular or extracellular loops. Regions with a high amount of basic residues are marked by +; − indicates a negatively charged membrane. B, Principle of “rescue of protein function” experiments: coexpression of α1A-trc or spdot-trc with the tail construct (α1–iD–TM4). The location of an alternative splice site in the TM3–TM4 loop of the α1 subunit is marked by a gray rectangle. C, Sequence alignment at the amino acid and nucleotide level of α1A wild-type, oscillator (spdot-trc), and the splice variant spdot-elg at amino acid positions 306–330 of the N-terminal region of the TM3–TM4 loop. The oscillator mutation (spdot) leads to a frame shift following TM3, running into an early STOP codon within the eight additional amino acid spanning cassette that is present in α1A wt. This results in spdot-trc if only one of the two splice acceptor sites is used or in spdot-elg with usage of both splice acceptor sites. The arrow points to the corresponding amino acid position of the STOP codon within the α1A wt sequence. D, Various truncated constructs. The α1A-trc variant is truncated at an amino acid position equivalent to the STOP codon in spdot-trc. In spdot-trc(RRKRRH), the positively charged motif RRKRRH is introduced into the oscillator variant. In α1A-trc(GDIT), the positively charged motif is exchanged for the corresponding amino acids from spdot. The mutants α1A-trc(T309R), spdot-trc(R309T), and spdot-trc(L308S/T309R) represent amino acid exchanges at the end of TM3, introducing elements from the wild-type α1 or spdot. E, Complementation constructs for determination of domains important for ion channel function. Bars represent parts of the mature protein (amino acid positions 29–424). Black boxes are the transmembrane domains, and the hatched box corresponds to the myc epitope used for detection of the construct [either at the N terminus (myc–α1–iD–TM4) or at the C terminus (α1–iD–TM4-myc)]. Constructs α1–iDΔ19–TM4, α1–iDΔ37–TM4, and α1–iDΔ62–TM4 are truncated at the N terminus of the tail construct; numbers of amino acids deleted are indicated.
Site-directed mutagenesis.
For generation of single point mutations [α1A(R309T), α1B(R309T), α1A-trc(R309T), spdot-trc(T309R), and spdot-trc(L308S/T309R)] or domain exchanges [α1A-trc(GDIT), spdot-trc(RRKRRH), myc–α1–iD–TM4, α1–iD–TM4–myc, pc–α1–iD–TM4–myc, and α1–iD–TM4–myc, with “pc” indicating positively charged], an overlap extension PCR strategy was used (Fig. 1). An overlap extension PCR requires two rounds of PCR. Every PCR was set up as follows: 100 ng of template DNA; 10 mm each dATP, dCTP, dGTP, and dTTP; 100 pmol of each sense and antisense primer and 2 units of high-fidelity Taq polymerase (Roche) in supplied polymerase buffer. PCR conditions were 5 min at 95°C, 5 min at 52°C, and 5 min at 72°C in the first cycle, followed by 28 cycles with 1 min at 95°C, 2 min at 52°C, and 3 min at 72°C. The last cycle ended with a 10 min 72°C amplification step. The first PCR reactions were done using a mutated sense primer combined with a parental antisense primer and a mutated antisense primer with a parental sense primer. In a second round of PCR, both mutated fragments were used as templates and submitted to two cycles without primers to allow efficient annealing within the overlapping region and elongation at the 3′ ends of the amplimers by the Taq polymerase. Finally, both parental primers were added and the same PCR conditions were used as shown above for amplification of the final PCR fragments. PCR fragments were cut with restriction enzymes as close as possible to the mutated site to minimize PCR-generated sequence and reinserted into GlyRα1. All mutated clones were sequenced across the PCR-generated sequence to verify successful mutagenesis using an ABI Sequencer (Applied Biosystems).
Spinal cord neuronal culture and genotyping.
Primary motoneuron cultures were prepared at embryonic day 12 (E12) following a modified protocol by Hoch et al. (1989). Briefly, every single spinal cord was trypsinized using 1 ml of trypsin/EDTA (1 mg/ml) and 10 μl of DNase I (final concentration 0.1 mg/ml), incubating the suspension at 37°C for 30 min. Trypsinization was stopped with 100 μl of fetal calf serum (final concentration, 10%). After a three-step trituration, the cells were centrifuged at 125 × g for 10 min. Trituration was repeated. Cells were plated to a density of 500,000 cells/3 cm dish and incubated at 37°C, 5% CO2, 95% humidity. A fast decomposition protocol was used for DNA extraction from the embryos' tails (Brodbeck et al., 2002). In brief, tails were incubated with buffer A (25 mm NaOH, 0.2 mm Na2EDTA) and heated at 95°C for 30 min while shaking. After cooling to 4°C, 75 μl of 40 mm Tris-HCl, pH 7.0, was added. Using 1.5 μl of tail DNA, a PCR was set up as follows: 4 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 63°C, and 30 s at 72°C, and a long elongation step (10 min at 72°C) was included after the last cycle. Primers used for amplification were mma1–sin7 (5′-GCCTCCGTGCTTTCTCCCTGC-3′) and mma1–ain8 (5′-CCCAGCCACGCCCCAAAG-3′).
Recombinant adeno-associated viral vectors.
To allow an efficient delivery of the C-terminal tail constructs into spinal cord neurons, a recombinant adeno-associated viral (rAAV) vector was used for expression of the tail construct. Therefore, three different tail variants (myc–α1–iD–TM4, pc–α1–iD–TM4–myc, and α1–iD–TM4) were cloned into a rAAV plasmid generating rAAV–6P–Cminibi.myc–α1–iD–TM4.Ven, rAAV–6P–Cminibi.pc–α1–iD–TM4–myc.Ven, and rAAV–6P–Cminibi.α1–iD–TM4.Ven. The virus was produced in HEK293 cells after transfection using the calcium-phosphate precipitation method. Purification of the virus was done using a modified protocol from Zolotukhin et al. (2002). Briefly, HEK293 cells were lysed 72 h after transfection. After lysis, the lysate was loaded on an iodixanol gradient (6–54%). Samples were ultracentrifuged, and 2 ml from the 40% iodixanol step was injected into a sterile tube and supplied to Q-column purification (HiTrap Q-HP; GE Healthcare). The eluted virus was concentrated using Amicon Ultra-15 Centrifugal Filter (Millipore).
Transfection/infection.
COS7 cells were transfected (1 μg plasmid/3 cm dish) using DEAE-Dextran (10 mg/ml). Cells were washed 30 min after transfection with DMEM and provided with 15 μl of chloroquin for 2 additional hours. After a second washing step, cells were incubated for 48 h before use in immunocytochemical experiments. HEK293 cells were transiently transfected (10 μg of plasmid/10 cm dish or 1 μg of DNA/3 cm dish) using the calcium-phosphate precipitation method. Phosphorylated enhanced GFP was cotransfected to control for transfection efficiency. For rescue experiments, the tail construct was used in excess compared with the trc variant (1:9; 0.3 μg of DNA of trc/2.7 μg of appropriate tail). All experiments concerning immunocytochemistry, protein biochemistry, and electrophysiological recordings were performed 48 h after transfection. Spinal cord neurons were transfected at 9 d in vitro (DIV) with the appropriate tail construct using Lipofectamine 2000 according to the instructions of the manufacturer (Invitrogen). Infection of spinal cord neurons/2 μl of virus (1:1) of rAAV–6P–Cminibi.myc–α1–iD–TM4.Ven and 1 μl of helper virus rAAV–hSyn–tTA (Wallace et al., 2008) were used for infection of spinal cord neurons at DIV9. Cells were used for staining 14–21 d after infection.
Protein biochemistry.
For membrane protein analysis, crude cell membranes were prepared from transfected cells as described by Sontheimer et al. (1989). For detection of the GlyRα variants, the monoclonal antibody pan-α (Mab4a, 1:200 dilution) was used, and for GlyRα1, the specific α1-antibody (Mab2b, 1:500 dilution) was used. Myc-tagged tail constructs were detected by either monoclonal or polyclonal myc antibodies (9E10) in a 1:200 dilution (Santa Cruz Biotechnology). As secondary antibodies, gαmIgG or gαrIgG coupled to cyanine 5 (Cy5) (1:250) were used (Dianova). Proteins were visualized with the fluoroimager STORM 860 (GE Healthcare).
Biotinylation assay.
Assays were performed with transfected COS7 cells 48 h after transfection as described previously (Oertel et al., 2007).
[3H]strychnine binding test.
Binding of [3H]strychnine (PerkinElmer Life and Analytical Sciences) (specific activity, 23.7 Ci/mmol) to crude membrane fractions was determined using a filtration assay (Hoch et al., 1989) with concentrations of free ligand ranging up to 200 nm. Binding data were analyzed using the Origin 7.0 program.
Immunocytochemistry.
Transfected cells were fixed using paraformaldehyde (4%) and sucrose (4%) in PBS (1.5 mm KH2PO4, 6.5 mm Na2HPO4, 3 mm KCl, and 137 mm NaCl) for 10 min at room temperature. After blocking in PBS with 5% goat serum for 30 min at room temperature, cells were incubated with the primary antibody (pan-α, anti-α1, or anti-myc) for 1 h at room temperature, followed by three washing steps in PBS. Afterward, the secondary antibody (1:200) was supplied for an additional hour at room temperature (gαmIgG–Cy3, gαmIgG–dichlorotriazinylamino-fluorescein (DTAF), or gαrIgG–DTAF; Dianova). For permeabilization of cells, 0.1% Triton X-100 was used during the blocking procedure. Slides were analyzed using a confocal microscope (Leica). For live stain of cells, transfected cells were incubated with the primary antibody (1:25) applied into the cell culture medium for 1 h at 4°C before fixation. In experiments using HEK293 cells, GFP and untransfected cells were used as negative controls as well as the secondary antibodies alone (data not shown). Neurofilament staining was performed as a positive control in spinal cord neuronal cultures, and again the secondary antibodies alone were used as negative controls (data not shown).
Digital image acquisition, processing, and analysis.
Images were 8-bit-encoded digital images acquired on a Leica TCS SP2 AOBS confocal microscope. Live cell imaging was performed with a micro Live Cell Incubation System (H. Saur). To avoid crosstalk or bleed through in dual and multiple color imaging, a sequential scanning method was used so that all the emission spectra are sufficiently separated from each other at the initial step of image acquisition. The sequential acquisition method was performed by exciting only one fluorophore at a time and detecting within the range of the emission spectra of the fluorophore concomitantly. For the purpose of colocalization analysis, confocal images were acquired with an aqueous immersion objective (63×, numerical aperture 1.2) in live cell studies. A sequential acquisition method was usually applied to unambiguously distinguish the different emission spectra of different fluorophores in the sample. Selected dual- or triple-color composite images were analyzed with the Leica Confocal Software for colocalization of different fluorophores. This digital analysis of colocalization of two or more fluorescent molecules provides information as to whether the fluorescence signals occupy the same pixel in the analyzed image. Compared with commonly used “visualized colocalization” by simple overlapping, the digital colocalization analysis presents precise spatial localization and accurate fluorescence intensities of individual fluorescent molecules at each pixel in the entire field of recording. The colocalization measurement performed in this study is based on a statistical approach that performs intensity correlation coefficient-based analysis.
mRNA calculation.
Transcript analysis was performed using the software NIH ImageJ. The amount of transcript was calculated from at least four independent experiments. As a control, we used β-actin as a housekeeping gene. The normalized transcript amount was calculated as the ratio of the amount of transcript of interest and the amount of transcript found for β-actin (trabs/β-actinabs).
Electrophysiological recordings.
Membrane currents were measured applying the patch-clamp technique in the whole-cell recording configuration. Current signals were amplified with an EPC-7 or EPC-9 amplifier (HEKA). Whole-cell recordings from transfected HEK293 cells were performed by application of ligand (glycine, in μm: 100, 500, or 3000) or coapplication of glycine and the antagonist strychnine (10 μm) using a U-tube, bathing the suspended cell in a laminar flow of solution, giving a time resolution for equilibration of 10–30 ms. The external buffer consisted of the following (in mm): 137 NaCl, 5.4 KCl, 1.8 CaCl2, 1.0 MgCl2, and 5.0 HEPES, pH adjusted to 7.2 with NaOH. The internal buffer contained the following (in mm): 120 CsCl, 20 N(Et)4Cl, 1.0 CaCl2, 2.0 MgCl2, 11.0 EGTA, and 10.0 HEPES, pH adjusted to 7.2 with CsOH. Current responses were measured at a holding potential of −60 mV. All experiments were performed at room temperature (∼22°C). Recording pipettes were fabricated from borosilicate capillaries with open resistances of 5–6 MΩ.
Results
Generation of α1A-trc and C-terminal tail variants
The phenotype of the Glra1 mouse mutant oscillator is attributed to a complete loss of the glycine receptor isoform GlyRA, caused by the absence of the ligand binding subunit variant α1. The overall topology of a full-length α1 subunit is determined by four TMs connected with intracellular or extracellular loops, an extracellular N terminus, and a short extracellular C terminus (Fig. 1). Compared with the α1 wild-type subunit sequence, the oscillator allele of Glra1 represents a 9 bp microdeletion, followed by a 2 bp insertion. Depending on the differential use of two splice acceptor sites in exon 9, two transcripts are derived from Glra1 in the wild type: transcript α1A carries an insertion of 8 aa to the TM3–TM4 loop, whereas transcript B lacks this insert (Malosio et al., 1991). Accordingly, two distinct transcripts originate from the mutant allele Glra1spd-ot. Inclusion of additional 24 nt by alternate splice acceptor selection in exon 9 results in transcript spdot-trc (Fig. 1). This transcript encodes a frame shift of 19 amino acid positions followed by an early STOP codon, truncating the cytoplasmic TM3–TM4 loop at its N-terminal end. In contrast, exclusion of the 24 bp insert by use of the downstream splice acceptor sites produces the mature transcript spdot-elg, which predicts a 150 amino acid frame-shift sequence following the site of the deletion (Buckwalter et al., 1994).
During heterologous expression in HEK293 cells, none of the Glra1spd-ot splice variants formed functional GlyR channels (Fig. 2). Given the high degree of heterogeneity of TM3–TM4 loop sequences among Cys-loop receptors (Breitinger and Becker, 2002), we hypothesized that these segments may not be required for folding of the highly conserved TM regions, including TM4. Assuming that TM4 represents an independently folding domain, we postulated that a C-terminal peptide construct covering most of the iD and TM4 (α1–iD–TM4) might complement the channel function of the TM4 truncated α1 mutant. To separate effects of subunit truncation from those of the oscillator frame shift, the truncated construct α1A-trc was generated, encoding the N-terminal 330 amino acid residues of wild-type α1A (Fig. 1B,C). The amino acid residues between positions 308 and 330 are localized C-terminally to TM3. To assess the influence of the wild-type TM3–TM4 loop sequence on TM integration of the nascent receptor subunit (Fig. 1B), N-terminal truncations of the α1–iD–TM4 construct were generated (Fig. 1D,E). The oscillator allele encodes an amino acid substitution starting at the cytoplasmic face of TM3: constructs spdot-trc(T309R) and spdot-trc(L308S/T309R) represent position-specific back mutations into the wild-type sequence. In constructs α1A(R309T) and α1A-trc(R309T), the corresponding oscillator amino acid positions were introduced into the wild-type sequences. The oscillator variant spdot-trc lacks the motif RRKRRH, a stretch of basic residues important for α1 subunit cell surface integration (Fig. 1C,D) (Sadtler et al., 2003). In the wild-type-derived α1A-trc construct α1A-trc(GDIT), this basic motif was mutated to corresponding oscillator positions and vice versa to wild-type amino acids in an oscillator sequence generating spdot-trc(RRKRRH) (Fig. 1D). The Glra1 C-terminal sequence used as an isolated tail construct for coexpression with truncated α1 variants harbors most of the TM3–TM4 loop, TM4, and the C terminus (α1–iD–TM4). A signal peptide and a N-terminal myc epitope were added to the tail sequence (myc–α1–iD–TM4) for easy immunodetection (Fig. 1E). Recordings from isolated expression of α1A-trc in HEK293 cells revealed that it is not able to generate functional Cl− channels, and neither are spdot-trc or spdot-elg (Fig. 2).
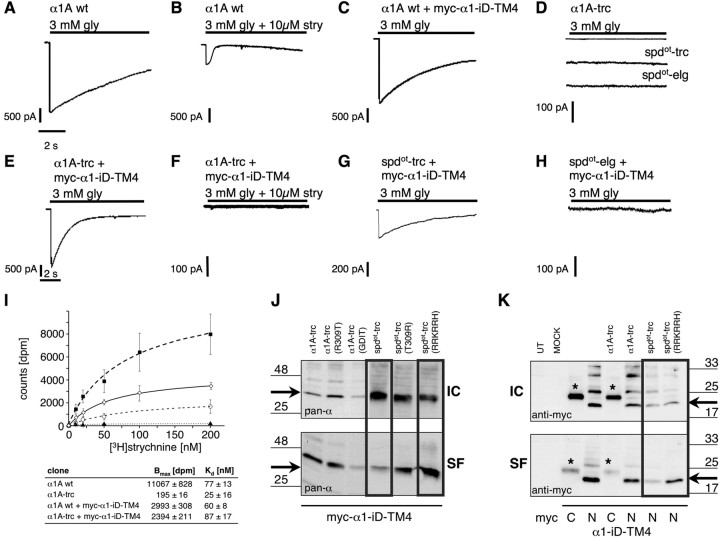
Figure 2.
Rescue of function experiments of spdot constructs compared with truncated α1 variants. Using whole-cell recordings from transfected HEK293 cells, maximal glycine (gly)-gated currents were determined with 3 mm gly: A, α1A wt; B, α1A wt blocked with strychnine (stry) (3 mm gly + 10 μm stry); C, α1A wt plus myc–α1–iD–TM4; D, α1A-trc, spdot-trc, and spdot-elg; E, α1A-trc plus myc–α1–iD–TM4; F, same as in E but antagonized with strychnine; G, spdot-trc plus myc–α1–iD–TM4; H, spdot-elg plus myc–α1–iD–TM4. Recordings were performed at 21–23°C, −60 mV, [Cl−]in = [Cl−]out. Agonists were applied via a U-tube for 10 s. I, [3H]Strychnine binding to membrane preparations of HEK293 cells transfected with α1A wt (filled squares), α1A wt + myc–α1–iD–TM4 (open diamonds), α1A-trc (filled triangles), or α1trc together with myc–α1–iD–TM4 (open circles). Apparent affinities (Kd) and total number of binding sites (Bmax) are shown below. J, K, Plasma membrane integration of spdot variants depends on the number of positively charged residues behind TM3. Surface proteins were labeled using NHS-biotin after pull down with streptavidin-coupled agarose beads. Western blots show the amount of surface protein (SF) compared with intracellular nonlabeled protein (IC) using the pan-α GlyR antibody, Mab4a. GFP-transfected HEK293 cells were used as internal control (K; MOCK). J, Comparison of intracellular (IC) and surface (SF) protein of α1A-trc and spdot-trc variants coexpressed with myc–α1–iD–TM4. Arrows point to the appropriate molecular weight of 33 kDa of spdot-trc and truncated α1 variants. K, Surface integration analysis of the N- and C-terminally myc-tagged α1–iD–TM4 variants. Untransfected (UT) and GFP-transfected HEK293 cells (MOCK) served as internal controls. Cotransfection of myc–α1–iD–TM4 (“N”) with spdot-trc(RRKRRH) led to a higher amount of surface-integrated tail protein compared with spdot-trc without the basic motif (black box). Black arrows point to the appropriate molecular weight of 20 kDa for the myc–α1–iD–TM4. * marks the C-myc-tagged α1–iD–TM4 (“C”), which always runs at a higher molecular weight of ∼25 kDa.
Rescue of the oscillator ion channel function
After transfection of α1A wt or α1B wt into HEK293 cells, functional Cl− channels were observed that could be antagonized with strychnine (Fig. 2A,B, Table 1). Coexpression of α1A wt together with the tail domain in a 1:1 ratio (myc–α1–iD–TM4) had no significant influence on glycine-evoked α1 currents of wild-type subunits (Fig. 2C, Table 1). During isolated expression in HEK293 cells, recombinant oscillator constructs were functionally inactive (Fig. 2D). In contrast, coexpression of the constructs myc–α1–iD–TM4 with α1A-trc or spdot-trc rescued glycine-gated ion channels (Fig. 2E,G). The glycinergic Cl− current of the complemented receptor complex was sensitive to the GlyR antagonist strychnine (Fig. 2F). Likewise, the short oscillator variant spdot-trc was rescued, yet with lower efficiency (Fig. 2G). In contrast, the elongated oscillator variant spdot-elg was not rescued (Fig. 2H, Table 1). This may be attributed to the long intracellular missense sequence of spdot-elg that may hinder an interaction with the construct myc–α1–iD–TM4. As obtained from glycine displaceable [3H]strychnine binding studies on membrane preparations from transfected HEK293 cells, the total number of antagonist binding sites (Bmax) was highly decreased in samples of truncated α1 subunit protein, reaching only 1.8% of α1A wild-type levels (Fig. 2I). Coexpression of α1A wt together with myc–α1–iD–TM4 lowered the total number of binding sites (Bmax = 2993 ± 308 dpm), most likely attributable to the transfected molar ratio of 1:1 (α1A wt/myc–α1–iD–TM4), resulting in an effective dilution of DNA coding for the active binding site. The Kd value was essentially unaffected compared with the isolated wild type (Fig. 2I). Coexpression of α1A-trc together with myc–α1–iD–TM4 raised the Bmax to ∼50% of wild type plus myc–α1–iD–TM4 corresponding to 22% of the isolated wild type. Binding affinities were similar for α1 wild-type and coexpression of α1A-trc with myc–α1–iD–TM4 (Fig. 2I). The complementation of GlyR ion channel function by independent coexpression of N-terminal and C-terminal α1 subunit fragments suggested that this GlyR subunit harbors independently folding domains.
Table 1.
Whole-cell maximal currents [Imax] for various coexpressed GlyR domains
| Clone | n | Imax (pA) 3 mm gly | n | Imax (pA) 500 μm gly |
|---|---|---|---|---|
| α1B wt | 4 | 1885 ± 947 | 2 | 2518 ± 1831 |
| α1A wt | 15 | 1826 ± 486 | 16 | 3040 ± 591 |
| α1A wt + myc–α1–iD–TM4 | 10 | 1196 ± 296 | 10 | 1977 ± 514 |
| α1A wt + α1–iD–TM4 | 6 | 1061 ± 228 | ||
| α1B(R309T) | 5 | 921 ± 648 | ||
| α1A(R309T) | 12 | 2376 ± 602 | ||
| α1A-trc | 8 | 0 ± 0 | ||
| α1-iD-TM4 | 6 | 0 ± 0 | ||
| α1A-trc + myc–α1–iD–TM4 | 37 | 1078 ± 240 | 20 | 1433 ± 218 |
| α1A-trc(R309T) | 8 | 0 ± 0 | ||
| α1A-trc(R309T) + myc–α1–iD–TM4 | 9 | 171 ± 44 | 4 | 0 ± 0 |
| α1A-trc(GDIT) | 3 | 0 ± 0 | ||
| α1A-trc(GDIT) + myc–α1–iD–TM4 | 9 | 469 ± 159 | 5 | 636 ± 371 |
| spdot-trc | 4 | 0 ± 0 | ||
| spdot-elg | 6 | 3 ± 3 (1/6)* | ||
| spdot-trc + spdot-elg | 6 | 0 ± 0 | ||
| spdot-trc + myc–α1–iD–TM4 | 19 | 182 ± 85 | 17 | 353 ± 88 |
| spdot-elg + myc–α1–iD–TM4 | 8 | 16 ± 11 | ||
| spdot-trc(T309R) | 6 | 0 ± 0 | ||
| spdot-trc(T309R/L308S) | 6 | 0 ± 0 | ||
| spdot-trc(RRKRRH) | 6 | 5 ± 5 (1/6)* | 6 | 0 ± 0 |
| spdot-trc(T309R) + myc–α1–iD–TM4 | 6 | 63 ± 14 | ||
| spdot-trc(T309R/L308S) + myc–α1–iD–TM4 | 4 | 83 ± 48 | ||
| spdot-trc(RRKRRH) + myc–α1–iD–TM4 | 6 | 2281 ± 708 | 6 | 3128 ± 946 |
HEK293 cells expressing different GlyR variants without or with coexpressed tail domains were patched after 48 h after tranfection; n indicates number of cells recorded; two different agonist concentrations were applied (500 μm and 3 mm glycine);
* indicates that only one cell responded. According to two splice acceptor sites in exon 9 of Glra1, α1A wt, and α1B wt (B, deletion; A, insertion; compare also Fig. 1) were generated for in vitro studies presenting the two α1 wild-type proteins existing in mice resulting from alternatively spliced α1 transcripts.
Frame-shift positions within TM3–TM4 loop harbor functionally important sites
Although constructs α1A-trc and spdot-trc both were rescued by a tail domain (α1–iD–TM4), they differed in rescue efficiency. The variant spdot-trc carries a C-terminal stretch of 19 missense amino acids (Fig. 1C). The corresponding wild-type sequence contains a basic motif involved in surface integration and G-protein βγ binding (Sadtler et al., 2003; Yevenes et al., 2006). As evident from human hyperekplexia mutations, the cytosolic arginine residues flanking transmembrane regions 2 and 4 are important for GlyR biogenensis and channel function (Langosch et al., 1993; Rea et al., 2002). The cytosolic position R309, flanking TM3, is mutated in oscillator (Buckwalter et al., 1994) and possibly contributes to protein dysfunction. Incorporation into the plasma membrane of oscillator polypeptides carrying either arginine 309 [spdot-trc(T309R)] or the entire basic motif [spdot-trc(RRKRRH)] was analyzed using surface protein biotinylation. In COS7 cell transfections, most of the wild-type α1 protein was labeled, indicating that it was exposed to the extracellular surface, whereas spdot-trc constructs accumulated in intracellular fractions (Fig. 2J). Indeed, the insertion of either one of the basic motifs into the stretch following TM3 significantly increased the surface protein fraction (Fig. 2J). In contrast, introduction of corresponding oscillator residues (T309 and GDIT319–322) into the wild-type sequence of α1A-trc variants reduced surface accumulation and shifted to the intracellular compartment or loss of antigen, respectively (Fig. 2J). These observations suggest that residues R309 and RRKRRH318–323 enhance plasma membrane integration. In addition, motif RRKRRH contributed to the interactions with the myc–α1–iD–TM4 construct, because the amount of surface-integrated myc–α1–iD–TM4 rose during inclusion of positively charged amino acids downstream of the oscillator mutation site (Fig. 2K).
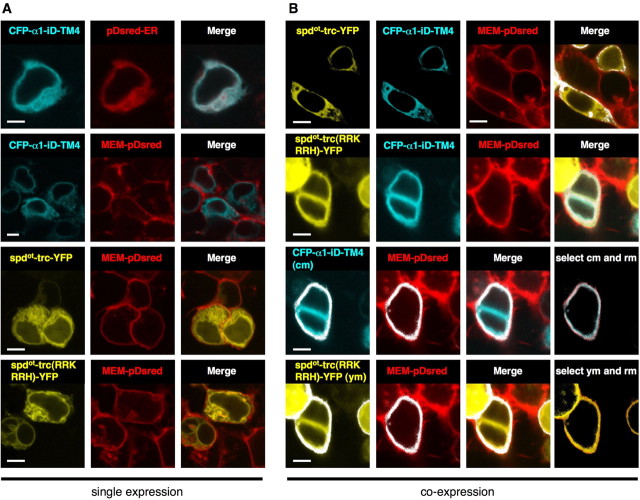
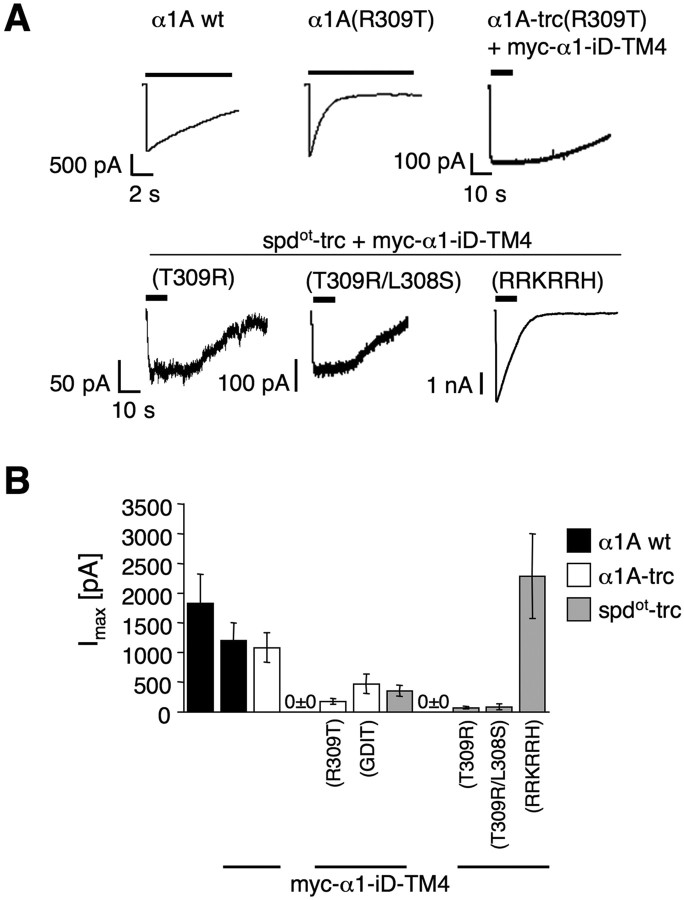
The synergism of the truncated variants and the tail construct α1–iD–TM4 was analyzed by tagging both with fluorescent proteins. During isolated expression of the yellow fluorescent protein (YFP)-tagged constructs, spdot-trc and spdot-trc(RRKRRH), fluorescence in the cell membrane was low (Fig. 3A). In contrast, coexpression with a cyan fluorescent protein (CFP)-tagged α1–iD–TM4 domain construct enforced the appearance of both spdot subunit variants at the cell surface, as evident from colocalization with the plasma membrane marker MEM–Discosoma red (DsRed)–monomer (Fig. 3B) (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). Apparently, the presence of both domains targeted the tagged polypeptides to the plasma membrane. In contrast, isolated expression resulted in intracellular accumulation consistent with a synergistic effect between both GlyR domains in ion channel formation. In electrophysiological recordings from transfected HEK293 cells, coexpression of spdot-trc(RRKRRH) and myc–α1–iD–TM4 generated wild-type levels (α1A wt and α1A wt + myc–α1–iD–TM4) of glycine-evoked maximal currents (Fig. 4, Table 1). The amino acid substitution of R309T in α1A wild type had no effect on Imax values [similar results for α1B(R309T) (Table 1)] but accelerated desensitization compared with the α1A wt (Fig. 4A). In α1A-trc substitution, R309T decreased rescue efficiency of the myc–α1–iD–TM4, coinciding with a loss of α1A-trc(R309T) from the plasma membrane (Fig. 4A,B). A similar reduction in Imax values was observed for coexpression of α1A-trc(GDIT) with myc–α1–iD–TM4 compared with coexpression with α1A-trc (Fig. 4B, Table 1). Conversely, the basic arginine at position 309 in spdot-trc(T309R) enhanced membrane integration (Fig. 2J), with a rescue efficiency similar to spdot-trc (Fig. 4, Table 1). The double-mutant spdot-trc(T309R/L308) was also not able to enhance rescue efficiency (Fig. 4B). Rescue of ion channel function was most efficient when spdot-trc(RRKRRH) was coexpressed with myc–α1–iD–TM4, reconstituting ion channel function to wild-type levels. Loss of the basic stretch RRKRRH was the most significant pathological exchange in the oscillator subunit variant, and its reversal the most efficient way to rescue GlyR channels (Fig. 4A,B, Table 1).
Figure 3.
CFP–α1–iD–TM4 and spdot-trc(RRKRRH)–YFP are cooperatively transported to the plasma membrane. A, Top two lanes, HEK 293 cells were cotransfected with CFP–α1–iD–TM4 and pDsRed–ER or MEM–DsRed–monomer, merged images (right picture). Note that there is hardly any colocalization between CFP–α1–iD–TM4 and MEM–DsRed–monomer. A, Bottom two lanes, Cells cotransfected with spdot-trc–YFP and MEM–DsRed–monomer or spdot-trc(RRKRRH)–YFP and MEM–DsRed–monomer. Right pictures represent the merged images. Note that the amount of plasma membrane protein is small when the single domains CFP–α1–iD–TM4, spdot-trc–YFP, or spdot-trc(RRKRRH)–YFP are transfected (right pictures, respectively). B, Coexpression of the tail variant CFP–α1–iD–TM4 together with trc variants resulted in an increase in surface protein, merged pictures of top two lanes in B. Coexpression of CFP–α1–iD–TM4, spdot-trc–YFP, and MEM–DsRed–monomer. B, Top lane, Coexpression of spdot-trc(RRKRRH)–YFP, CFP–α1–iD–TM4, and MEM–DsRed–monomer. B, Second lane, Colocalization analysis of coexpression of constructs used in the top two lanes of B: masked images of selected dots from both cyan (cm) and red (rm) signals of approximately same intensity values (shown as white dots in the images), overlay of selected cm and rm. Right picture, Third lane in B. Fourth lane, Colocalization analysis: same method as shown in third lane, yellow channel (ym), red channel (rm); overlay of ym and rm; right picture, bottom lane in B. Scale bars, 4 μm.
Figure 4.
Effects of arginine residues within the TM3–TM4 loop on physiological properties. A, Representative traces of glycine-gated currents (3 mm) recorded from HEK293 cells transfected with either α1A wt or α1A(R309T). Truncated variants α1A-trc or spdot-trc, α1A-trc(R309T), spdot-trc(T309R), spdot-trc(T309R/L308S), or spdot-trc(RRKRRH) were cotransfected with myc–α1–iD–TM4. B, Bar diagram representing maximal currents Imax ± SEM (in picoamperes) compared with wild type, n = 8–24 cells measured in at least three different batches of transfected cells.
The “tail” domain provides structural signals for GlyR channel rescue
After the identification of the basic motif RRKRRH as the structural determinant important for ion channel reconstitution from split subunit constructs, we attempted to define interacting motifs of the tail domain.
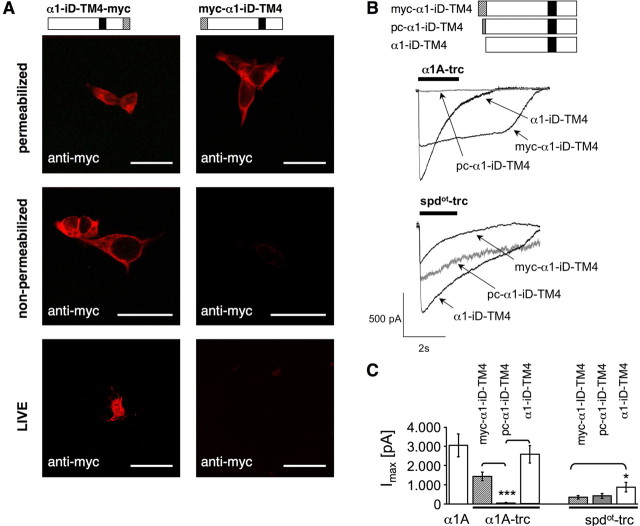
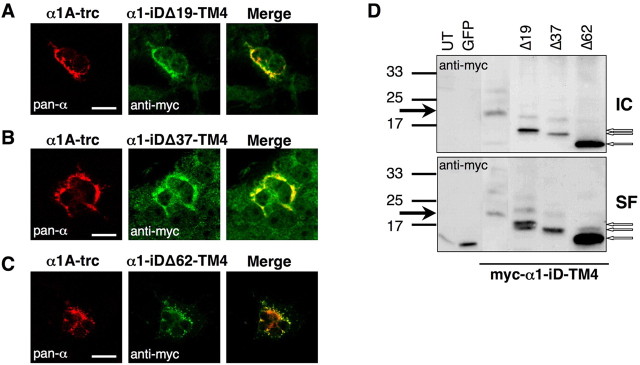
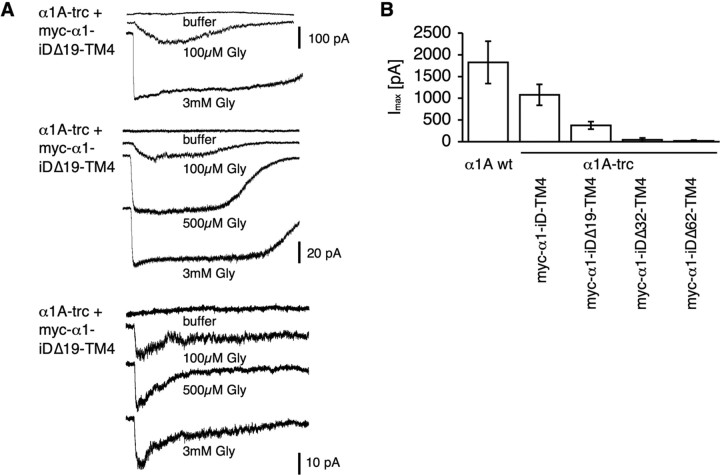
To determine the transmembrane orientation of isolated TM4 constructs, a myc epitope was attached at the C terminus of the tail domain construct (α1–iD–TM4-myc). Indeed, a comparison of permeabilized and nonpermeabilized cells revealed a canonical GlyR topology of the C-terminal tail domain (Figs. 1E, 5A). Thus, topogenic signals important for orientation of TM4 were not affected by the generation of the rescue tail variants. To analyze the influence of the myc epitope or the basic motif in front of the tail sequence on rescue efficiency, tail variants, e.g., pc–α1–iD–TM4-myc and α1–iD–TM4, were generated. Tails without the myc epitope coexpressed with either α1A-trc or spdot-trc doubled rescue efficiency (Fig. 5B,C, Table 2). The basic residues at the N terminus of the tail construct (pc–α1–iD–TM4) had no significant influence on spdot-trc. In contrast, in coexpression with α1A-trc harboring the RRKRRH motif on its own at the C-terminal end, Imax values were highly reduced (Fig. 5B,C, Table 2). The presence of the basic motif on both sites of the complementary GlyRα1 subunit constructs most likely did not allow a close vicinity of both GlyR domains necessary for rescue of ion channel function. Successive N-terminal truncations of the tail (constructs: α1–iDΔ19–TM4, α1–iDΔ37–TM4, and α1–iDΔ62–TM4) did not interfere with their surface expression in HEK293 cells (Fig. 6D). Coexpression with α1A-trc, however, demonstrated a loss of plasma membrane accumulation, consistent with a loss of cooperativeness between α1A-trc and the truncated tails (Fig. 6A–C). These observations were confirmed by whole-cell recordings, in which truncation of the tail domain in construct α1–iDΔ19–TM4 resulted in a dramatic decrease in the Imax value. With constructs α1–iDΔ37–TM4 or α1–iDΔ62–TM4, responses were virtually absent (Fig. 7, Table 3). Thus, the amino acid positions of the TM3–TM4 loop, which in the tail domain exist as a free N terminus, are essential for an interaction with α1A-trc to rescue ion channel function.
Figure 5.
Orientation and rescue efficiency of N-myc- and C-myc-tagged tail constructs. A, Bars schematically illustrate the tail variants myc–α1–iD–TM4 and α1–iD–TM4-myc, showing the location of the myc epitope (hatched box) and TM4 (black box). Transfected COS7 cells were used either unpermeabilized or permeabilized with 0.1% Triton X-100. Bottom pictures represent a live stain experiment. Live staining was performed using a 1 h incubation step at 4°C before fixation. Untransfected cells and GFP-transfected cells were used as controls (data not shown). Scale bars, 20 μm. B, The myc epitope was cut for determination of rescue efficiency by the tail sequence (α1–iD–TM4). Pc–α1–iD–TM4 is modified by harboring the basic stretch RRKRRH attached to the N terminus of the tail construct (gray box). Constructs lacking the myc epitope (α1–iD–TM4) were most efficient in rescue of the appropriate trc construct independent if they were of α1 or spdot-origin. C, Bar diagram of the calculated maximal current amplitudes. Note that, in spdot-trc + tail coexpressions, the maximal current amplitudes were doubled compared with spdot-trc + myc–α1–iD–TM4. *p < 0.01, ***p < 0.0001.
Table 2.
Rescue efficiency of α1–iD–TM4 sequence motifs
| Imax (pA) | Cell (#) | Glycine | |
|---|---|---|---|
| α1A wt | 3040 ± 591 | 16 | 3 mm |
| α1A-trc + myc–α1–iD–TM4 | 1433 ± 219 | 20 | 3 mm |
| α1A-trc + pc–α1–iD–TM4 | 37 ± 27 | 8 | 3 mm |
| α1A-trc + α1–iD–TM4 | 2553 ± 448 | 16 | 3 mm |
| spdot-trc + myc–α1–iD–TM4 | 352 ± 87 | 17 | 3 mm |
| spdot-trc + pc–α1–iD–TM4 | 418 ± 126 | 14 | 3 mm |
| spdot-trc + α1–iD–TM4 | 878 ± 243 | 7 | 3 mm |
Figure 6.
Subcellular localization of truncated tail variants α1–iDΔ19–TM4, α1–iDΔ37–TM4, and α1–iDΔ62–TM4. COS7 cells were transfected with truncated α1 (α1A-trc) and one of the shortened tails (A–C). Cells were permeabilized; for α1A-trc, the monoclonal antibody pan-α was used (red pictures). Tail variants were detected via their myc epitope (green pictures). Right panels represent merged pictures. D, Surface integration of truncated tail constructs without coexpression of α1A-trc compared with intracellular protein (IC). Untransfected (UT) and GFP-transfected HEK293 cells (MOCK) served as internal controls. Although high amounts of truncated tails were integrated into the plasma membrane (SF) when expressed alone (D), surface integration of the truncated tail variants decreased in coexpression with α1A-trc (A–C, right panels). Black arrows point to the appropriate molecular weight of 20 kDa for the myc–α1–iD–TM4. Truncated C-terminal tail proteins were marked with open arrows. Scale bars, 20 μm.
Figure 7.
Importance of the TM3–TM4 loop for rescue of protein function. A, Traces of glycine-gated (100 μm, 500 μm, and 3 mm) currents of truncated tails (α1–iDΔ19–TM4, α1–iDΔ37–TM4, or α1–iDΔ62–TM4) coexpressed with α1A-trc. B, Bar diagram demonstrating reduction in whole-cell maximal current amplitudes (in picoamperes) of tail variants coexpressed with α1A-trc compared with α1A wt.
Table 3.
Maximal currents of truncated α1–iD–TM4 variants
| Clone | n | Imax (pA) |
|---|---|---|
| α1A wt | 3 | 2059 ± 591 |
| α1A-trc + myc–α1–iD–TM4 | 14 | 1153 ± 260 |
| α1A-trc + myc–α1–iDΔ19–TM4 | 14 | 320 ± 76 |
| α1A-trc + myc–α1–iDΔ37–TM4 | 1 of 11 | 42* |
| α1A-trc + myc–α1–iDΔ62–TM4 | 1 of 8 | 21* |
*Indicates Imax of one cell responding of n cells total.
Rescue of endogenous oscillator α1 protein in primary neurons of ot/ot embryos
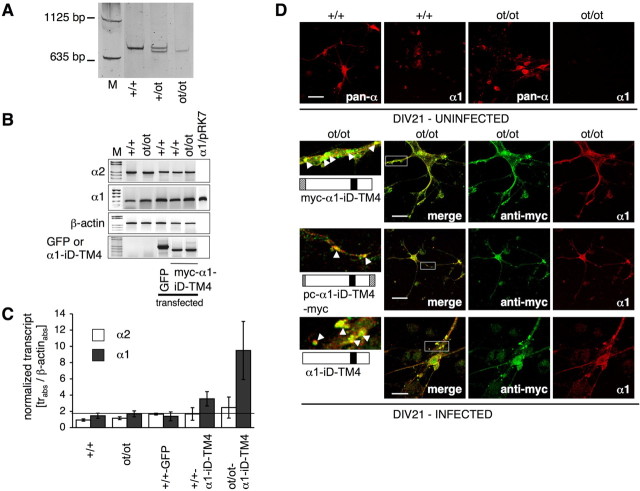
After a successful rescue of recombinant α1 subunit variants in heterologous HEK293 cell expression, we transferred this approach to the endogenous α1 subunit mutant of cultured spinal cord neurons from homozygous oscillator embryos. Spinal cord neurons were prepared from genotyped oscillator (ot/ot) and wild-type (+/+) embryos at E12 (Fig. 8A). After DIV7, the primary neurons were either transfected or virally infected with myc–α1–iD–TM4 construct, which was efficiently expressed at the mRNA level (Fig. 8B,C). Primers recognizing myc–α1–iD–TM4 did not detect the endogenous α1 transcript, and the primers designed to detect α1 transcripts did not interfere with detection of the exogenously expressed tail construct myc–α1–iD–TM4.
Figure 8.
Rescue of spdot protein by expression of myc–α1–iD–TM4 in spinal cord neuronal cultures from ot/ot mutant mice. Genotypes are as follows (A): +/+, homozygous wild-type; +/ot, heterozygous; and ot/ot, homozygous spdot. Cultured spinal cord neurons were transfected with myc–α1–iD–TM4 or a GFP control. B, After RT-PCR, transcript analysis was performed for endogenous GlyRα1 and GlyRα2, and GFP and myc–α1–iD–TM4 to analyze for transfection efficiency. Transcripts shown in C were normalized to β-actin; background level of α1 transcript was 1.9 (black line). In parallel, spinal cord neurons were infected with tail constructs encoded on a pAAV vector. D, Comparison of infected (DIV21; bottom) and uninfected spinal cord neurons isolated from either homozygous wild-type (+/+) or oscillator (ot/ot) mice (DIV21 uninfected). The α1-antibody Mab2b recognizes only the α1 variant, whereas pan-α recognizes all GlyRα variants. After infection with myc–α1–iD–TM4, pc–α1–iD–TM4-myc, or α1–iD–TM4, infected neurons expressed endogenous GlyRα1 protein (bottom 3 panels, right pictures, depicted in red), which colocalizes (merged pictures) with myc–α1–iD–TM4. White boxes give enlarged images on the left side; white arrowheads point to clusters of colocalized spdot and C-terminal tail proteins. Scale bars, 20 μm.
At the protein level, we could also observe an increase of spdot. The GlyRα1 subunit carries an N-terminal epitope that is recognized by the monoclonal α1 antibody (Mab2b). In both, native spinal cord (Kling et al., 1997) and primary neurons from homozygous oscillator mutants, the N-terminal epitope was absent, indicating a complete loss of the α1 subunit mutant in homozygous animals (Fig. 8D). Expression of the C-terminal construct, which is not recognized by the anti-α1 antibody, in ot/ot neurons, resulted in the appearance of N-terminal GlyRα1 antigen (Fig. 8D). The myc–α1–iD–TM4 construct was sorted into the same compartments as spdot, appearing to a lesser extent in the soma compared with the much higher amount in clusters localized at the dendrites of infected neurons (Fig. 8D, bottom lane, left pictures). The appearance in dendritic clusters of ot/ot neurons of endogenous α1 antigen was also observed for the other tail variants, e.g., pc–α1–iD–TM4 and α1–iD–TM4 (Fig. 8D). As shown above, these were able to rescue ion channel function when coexpressed with spdot-trc in HEK293 cells (Fig. 5, Table 2).
Our observations indicate that the oscillator mutant protein is rescued by coexpression of a C-terminal domain of the GlyRα1 subunit. This suggests that the C-terminal domain of the α1 subunit (α1–iD–TM4), comprising the TM3–TM4 loop, TM4, and the short C terminus, act as an independently folding domain of the GlyRα1 polypeptide. Moreover, the C-terminal tail domain is essential for rescue of the truncated ion channel function and, concomitantly, for trafficking of the spdot polypeptide toward the plasma membrane. Furthermore, transcript analysis revealed that the expression of the myc–α1–iD–TM4 construct associated with an increase in Glra1spd-ot transcripts, as detected by a primer combination detecting both the long and the short α1 splice variants but not transcripts of the exogenous expressed tail. In contrast, transcript levels of the GlyRα2 subunit variant were unaltered (Fig. 8B,C). Apparently, rescue of α1 subunit polypeptide has a positive feedback of α1 transcript levels, which as yet needs to be explained.
Discussion
Ion channel proteins of the Cys-loop receptor family share an overall transmembrane topology in which the large N-terminal domain (Breitinger et al., 2004) is followed by four transmembrane regions, with TM2 lining the ion pore. Here, we show that coexpression of C-terminally truncated α1 subunit variants together with a construct of the tail domain reconstituted GlyR function. Evidently, GlyRα subunits comprise two domains that, during expression as separate constructs, reconstitute the functionalities of a ligand-gated ion channel without being covalently attached to each other.
Structural implications of the oscillator mutation of the mouse
The functional role of the TM3–TM4 loop of GlyR subunits, which is less conserved among the subunit variants, is understood poorly. Mutant studies with the serotonin receptor subunit 5-HT3A and the GABAC receptor subunit ρ1 show that the TM3–TM4 loop can be replaced by a short linker without abolishing the fundamental function of ligand-gated ion conductance (Jansen et al., 2008). Nevertheless, the TM3–TM4 loop may be a modifier of ion channel function: splice variants of the GlyR subunits α1 and α3, which differ in their TM3–TM4 loop sequences, differ in desensitization behavior (Nikolic et al., 1998). Moreover, GlyR subunit variants share consensus sequences within the TM3–TM4 loop that are indicative of a functional specialization.
During receptor biogenesis and intracellular trafficking, structural motifs are thought to guide the spatial arrangement of the transmembrane domains and the TM3–TM4 loop within the individual subunit, as well as within the GlyR multi-subunit protein complex. In particular, a basic motif within the TM3–TM4 loop has been shown to be indispensable for subunit trafficking to the plasma membrane (Sadtler et al., 2003). Consistent with the “positive inside” rule, this motif has been postulated to compensate for the presence of positive charges in the M2–M3 ectodomain, which otherwise might interfere with a proper membrane integration of the M3 segment (Sadtler et al., 2003).
In the CNS of oscillator mice, two distinct α1 splice variants, i.e., spdot-trc and spdot-elg, are formed. The oscillator mutation of Glra1 not only cuts off a significant part of the α1 subunit sequence but also destroys motifs important for receptor biogenesis (Buckwalter et al., 1994; Sadtler et al., 2003). The corresponding mutant polypeptides spdot-trc and spdot-elg are virtually absent from the CNS of homozygous mutants, indicating that subunit stability is lost (Kling et al., 1997).
GlyR complementation and the basic motif within the TM3–TM4 loop
Efficiency of rescue by coexpression with the tail domain differed between the truncated α1 mutant, spdot-trc, and the elongated α1 variant, spdot-elg. This may be attributed to a potential sterical hindrance exerted by the long missense C terminus of spdot-elg. Although there are no folding models of the TM3–TM4 loops of Cys-loop receptors (Kukhtina et al., 2006), it may be speculated that the aberrant C-terminal domain of spdot-elg does not allow a close association with the tail domain or the negatively charged plasma membrane.
The basic motif RRKRRH, located within the TM3–TM4 loop of the wild-type α1 subunit, is affected by the oscillator frame shift, leaving only two basic residues within the frame-shift sequence of spdot-trc. A back mutation that reestablished the motif RRKRRH within the mutant sequence of spdot-trc dramatically increased surface expression of the mutant subunit spdot-trc(RRKRRH). Coexpression of the oscillator mutant construct spdot-trc(RRKRRH) with the tail domain (α1–iD–TM4) also increased the surface expression of the tail peptide. With the basic motif present, coexpression of both components also raised glycine-induced maximum currents to wild-type levels, indicating that these fragments had assembled into functional receptor channels. The basic motif also modulated the cell surface integration of the independently expressed α1–iD–TM4 domain: when expressed as isolated constructs, the α1–iD–TM4 peptides integrated efficiently into the cell surface, even when truncated on its N-terminal side (α1–iDΔ19–TM4, α1–iDΔ37–TM4, or α1–iDΔ62–TM4). When the tail α1–iD–TM4 was coexpressed with an N-terminal construct that lacked the basic motif, e.g., spdot-trc or α1A-trc(GDIT), surface integration of the tail α1–iD–TM4 was suppressed. In contrast, coexpression of α1–iD–TM4 together with α1A-trc, harboring the RRKRRH motif, promoted integration into the cell membrane. Although previous observations (Sadtler et al., 2003) indicate that the basic motif is important for cell surface integration of full-length α1 subunits, our data reveal that it also mediates protein–protein interaction during mutant receptor biogenesis. Accordingly, the motif may be involved in the intra-subunit interaction of distinct domains within the wild-type α1 receptor polypeptide. The promoting effect of the C-terminal tail domain on membrane integration and ion channel formation by α1 subunit mutants may be correlated with two major phases of protein biogenesis. (1) During protein oligomerization, the tail domain may serve as a structural component required for the receptor conformation that mediates cell surface integration and ion channel function. Thus, the basic motif may serve as a signal, bonding two separate polypeptide chains within a common protein fold. (2) In the endoplasmic reticulum (ER), the basic motif RRKRRH might serve as a retention signal (Holmes et al., 2002; Hawkins et al., 2004). Accordingly, the tail domain, α1–iD–TM4, would shield this signal and, thereby, promote surface integration. Indeed, homomerically expressed trc variants were retained in the ER compartment most likely attributable to the ER quality control system. This mechanism, however, is less likely because the mutant constructs spdot-trc and spdot-trc(R309T), lacking the basic motif, also retained in the ER.
Membrane orientation of the tail domain
Topogenic signals contribute to the membrane insertion of transmembrane domains. In bacterial membrane proteins, cytoplasmic domains contain more positive amino acid charges than periplasmic proteins do. Deduced from these observations, the positive inside rule also applies to eucaryotic proteins, although the tendency of positive charges to be absent from extracellular domains appears to be weaker. Accordingly, positive charges in connecting cytosolic loops may serve as topogenic signals for membrane integration (van Geest and Lolkema, 2000; Hessa et al., 2005). When the transmembrane orientation of the tail α1–iD–TM4 was analyzed, we consistently observed an orientation that corresponded to that of the wild-type α1 subunit. This orientation was not influenced by the position of the myc epitope included at either the N or the C terminus of the construct. Indeed, the presence of the positively charged amino acid residues at the cytoplasmic N terminus of the tail construct and its final orientation are consistent with the positive inside rule (Andersson et al., 1992). This also held true for the truncated variants α1–iDΔ19–TM4, α1–iDΔ37–TM4, and α1–iDΔ62–TM4, in which the positively charged motif 411RAKKIDKISR429 was oriented to the cytosol. In contrast, the rescue efficiency of tail constructs decreased with progressive N-terminal shortening of the tail domain, indicating that these sequences are involved in the interaction of the subunit fragments during ion channel assembly.
Upregulation of the endogenous mutant GlyRα1 by complementation
Rescue by α1 subunit tail constructs also applies to native neurons cultured from spinal cord of homozygous ot/ot mouse mutants. In ot/ot neurons, expression of the tail domains resulted in the rescue of the endogenous mutant α1 subunit and its correct sorting into the neuronal plasma membrane. This observation suggests that the α1–iD–TM4 domain acts as an independently folding domain in native spinal cord neurons.
Paradigmatic expression studies indicate that ligand-gated ion channels represent structural mosaics of elements derived from other precursor proteins (Villmann et al., 1997; Breitinger et al., 2004). Our experiments demonstrate that inhibitory GlyRs are composed of independently folding domains that assemble into a functional channel. Although the complementation of GlyR function reached levels of up to 50%, assembly of subunit fragments of the heteromeric NR1/NR2 receptor was reported for 10% functionality (Schorge and Colquhoun, 2003). This difference may be attributable to the structural characteristics of the GlyR and NMDA receptor complexes: although GlyRα1 subunits assemble into functional homopentamers, the tetrameric NMDA receptor complex requires a stepwise association process in which two NR1/NR2 heterodimers dimerize to assemble into the tetrameric receptor complex (Schüler et al., 2008; Villmann et al., 2008).
In summary, our observations show that the truncated subunit variant spdot-trc from oscillator is efficiently rescued by the C-terminal tail domain that was destroyed by the oscillator deletion. This complementation points to the importance of the TM3–TM4 loop for biosynthesis, clustering, and pharmacology of GlyRs and may have implications for novel approaches to gene therapy. Our data support the idea that the tail domain is able to independently interact with the remainder of the receptor protein. This conclusion is consistent with the hypothesis that GlyRs represent a class of ligand-gated channels composed of independent building blocks.
Footnotes
This work was supported by Deutsche Forschungsgemeinschaft Grants BE1138/5-6, STA399/9-1, and SFB636/A4, European Union (Neuro Cypress, HEALTH-F4-2008-202088), and Johannes und Frieda Marohn-Stiftung. Drs. Heike Meiselbach and Nima Melzer are gratefully acknowledged for support and helpful discussions. Rosa Weber and Annette Herold provided excellent technical assistance.
References
- Andersson H, Bakker E, von Heijne G. Different positively charged amino acids have similar effects on the topology of a polytopic transmembrane protein in Escherichia coli. J Biol Chem. 1992;267:1491–1495. [PubMed] [Google Scholar]
- Becker CM, Hoch W, Betz H. Glycine receptor heterogeneity in rat spinal cord during postnatal development. EMBO J. 1988;7:3717–3726. doi: 10.1002/j.1460-2075.1988.tb03255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker CM, Schmieden V, Tarroni P, Strasser U, Betz H. Isoform-selective deficit of glycine receptors in the mouse mutant spastic. Neuron. 1992;8:283–289. doi: 10.1016/0896-6273(92)90295-o. [DOI] [PubMed] [Google Scholar]
- Becker K, Hohoff C, Schmitt B, Christen HJ, Neubauer BA, Sandrieser T, Becker CM. Identification of the microdeletion breakpoint in a GLRA1null allele of Turkish hyperekplexia patients. Hum Mutat. 2006;27:1061–1062. doi: 10.1002/humu.9455. [DOI] [PubMed] [Google Scholar]
- Becker K, Breitinger HG, Humeny A, Meinck HM, Dietz B, Aksu F, Becker CM. The novel hyperekplexia allele GLRA1(S267N) affects the ethanol site of the glycine receptor. Eur J Hum Genet. 2008;16:223–228. doi: 10.1038/sj.ejhg.5201958. [DOI] [PubMed] [Google Scholar]
- Betz H, Laube B. Glycine receptors: recent insights into their structural organization and functional diversity. J Neurochem. 2006;97:1600–1610. doi: 10.1111/j.1471-4159.2006.03908.x. [DOI] [PubMed] [Google Scholar]
- Breitinger HG, Becker CM. The inhibitory glycine receptor - simple views of a complicated channel. Chembiochem. 2002;3:1042–1052. doi: 10.1002/1439-7633(20021104)3:11<1042::AID-CBIC1042>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Breitinger U, Breitinger HG, Bauer F, Fahmy K, Glockenhammer D, Becker CM. Conserved high affinity ligand binding and membrane association in the native and refolded extracellular domain of the human glycine receptor alpha1-subunit. J Biol Chem. 2004;279:1627–1636. doi: 10.1074/jbc.M303811200. [DOI] [PubMed] [Google Scholar]
- Brodbeck J, Davies A, Courtney JM, Meir A, Balaguero N, Canti C, Moss FJ, Page KM, Pratt WS, Hunt SP, Barclay J, Rees M, Dolphin AC. The ducky mutation in Cacna2d2 results in altered Purkinje cell morphology and is associated with the expression of a truncated alpha 2 delta-2 protein with abnormal function. J Biol Chem. 2002;277:7684–7693. doi: 10.1074/jbc.M109404200. [DOI] [PubMed] [Google Scholar]
- Buckwalter MS, Cook SA, Davisson MT, White WF, Camper SA. A frameshift mutation in the mouse alpha 1 glycine receptor gene (Glra1) results in progressive neurological symptoms and juvenile death. Hum Mol Genet. 1994;3:2025–2030. doi: 10.1093/hmg/3.11.2025. [DOI] [PubMed] [Google Scholar]
- Büsselberg D, Bischoff AM, Becker K, Becker CM, Richter DW. The respiratory rhythm in mutant oscillator mice. Neurosci Lett. 2001;316:99–102. doi: 10.1016/s0304-3940(01)02382-5. [DOI] [PubMed] [Google Scholar]
- Eulenburg V, Becker K, Gomeza J, Schmitt B, Becker CM, Betz H. Mutations within the human GLYT2 (SLC6A5) gene associated with hyperekplexia. Biochem Biophys Res Commun. 2006;348:400–405. doi: 10.1016/j.bbrc.2006.07.080. [DOI] [PubMed] [Google Scholar]
- Grudzinska J, Schemm R, Haeger S, Nicke A, Schmalzing G, Betz H, Laube B. The beta subunit determines the ligand binding properties of synaptic glycine receptors. Neuron. 2005;45:727–739. doi: 10.1016/j.neuron.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Harvey RJ, Depner UB, Wässle H, Ahmadi S, Heindl C, Reinold H, Smart TG, Harvey K, Schütz B, Abo-Salem OM, Zimmer A, Poisbeau P, Welzl H, Wolfer DP, Betz H, Zeilhofer HU, Müller U. GlyR alpha3: an essential target for spinal PGE2-mediated inflammatory pain sensitization. Science. 2004;304:884–887. doi: 10.1126/science.1094925. [DOI] [PubMed] [Google Scholar]
- Hawkins LM, Prybylowski K, Chang K, Moussan C, Stephenson FA, Wenthold RJ. Export from the endoplasmic reticulum of assembled N-methyl-d-aspartic acid receptors is controlled by a motif in the c terminus of the NR2 subunit. J Biol Chem. 2004;279:28903–28910. doi: 10.1074/jbc.M402599200. [DOI] [PubMed] [Google Scholar]
- Heinze L, Harvey RJ, Haverkamp S, Wässle H. Diversity of glycine receptors in the mouse retina: localization of the alpha4 subunit. J Comp Neurol. 2007;500:693–707. doi: 10.1002/cne.21201. [DOI] [PubMed] [Google Scholar]
- Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hoch W, Betz H, Becker CM. Primary cultures of mouse spinal cord express the neonatal isoform of the inhibitory glycine receptor. Neuron. 1989;3:339–348. doi: 10.1016/0896-6273(89)90258-4. [DOI] [PubMed] [Google Scholar]
- Holmes KD, Mattar PA, Marsh DR, Weaver LC, Dekaban GA. The N-methyl-d-aspartate receptor splice variant NR1–4 C-terminal domain. Deletion analysis and role in subcellular distribution. J Biol Chem. 2002;277:1457–1468. doi: 10.1074/jbc.M107809200. [DOI] [PubMed] [Google Scholar]
- Jansen M, Bali M, Akabas MH. Modular design of Cys-loop ligand-gated ion channels: functional 5-HT3 and GABA rho1 receptors lacking the large cytoplasmic M3M4 loop. J Gen Physiol. 2008;131:137–146. doi: 10.1085/jgp.200709896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsmore SF, Giros B, Suh D, Bieniarz M, Caron MG, Seldin MF. Glycine receptor beta-subunit gene mutation in spastic mouse associated with LINE-1 element insertion. Nat Genet. 1994;7:136–141. doi: 10.1038/ng0694-136. [DOI] [PubMed] [Google Scholar]
- Kling C, Koch M, Saul B, Becker CM. The frameshift mutation oscillator (Glra1(spd-ot)) produces a complete loss of glycine receptor alpha1-polypeptide in mouse central nervous system. Neuroscience. 1997;78:411–417. doi: 10.1016/s0306-4522(96)00567-2. [DOI] [PubMed] [Google Scholar]
- Kukhtina V, Kottwitz D, Strauss H, Heise B, Chebotareva N, Tsetlin V, Hucho F. Intracellular domain of nicotinic acetylcholine receptor: the importance of being unfolded. J Neurochem. 2006;97(Suppl 1):63–67. doi: 10.1111/j.1471-4159.2005.03468.x. [DOI] [PubMed] [Google Scholar]
- Langosch D, Herbold A, Schmieden V, Borman J, Kirsch J. Importance of Arg-219 for correct biogenesis of alpha 1 homooligomeric glycine receptors. FEBS Lett. 1993;336:540–544. doi: 10.1016/0014-5793(93)80872-r. [DOI] [PubMed] [Google Scholar]
- Lynch JW. Molecular structure and function of the glycine receptor chloride channel. Physiol Rev. 2004;84:1051–1095. doi: 10.1152/physrev.00042.2003. [DOI] [PubMed] [Google Scholar]
- Malosio ML, Grenningloh G, Kuhse J, Schmieden V, Schmitt B, Prior P, Betz H. Alternative splicing generates two variants of the alpha 1 subunit of the inhibitory glycine receptor. J Biol Chem. 1991;266:2048–2053. [PubMed] [Google Scholar]
- Mülhardt C, Fischer M, Gass P, Simon-Chazottes D, Guénet JL, Kuhse J, Betz H, Becker CM. The spastic mouse: aberrant splicing of glycine receptor beta subunit mRNA caused by intronic insertion of L1 element. Neuron. 1994;13:1003–1015. doi: 10.1016/0896-6273(94)90265-8. [DOI] [PubMed] [Google Scholar]
- Nikolic Z, Laube B, Weber RG, Lichter P, Kioschis P, Poustka A, Mülhardt C, Becker CM. The human glycine receptor subunit alpha3. Glra3 gene structure, chromosomal localization, and functional characterization of alternative transcripts. J Biol Chem. 1998;273:19708–19714. doi: 10.1074/jbc.273.31.19708. [DOI] [PubMed] [Google Scholar]
- Oertel J, Villmann C, Kettenmann H, Kirchhoff F, Becker CM. A novel glycine receptor beta subunit splice variant predicts an unorthodox transmembrane topology. Assembly into heteromeric receptor complexes. J Biol Chem. 2007;282:2798–2807. doi: 10.1074/jbc.M608941200. [DOI] [PubMed] [Google Scholar]
- Rea R, Tijssen MA, Herd C, Frants RR, Kullmann DM. Functional characterization of compound heterozygosity for GlyRalpha1 mutations in the startle disease hyperekplexia. Eur J Neurosci. 2002;16:186–196. doi: 10.1046/j.1460-9568.2002.02054.x. [DOI] [PubMed] [Google Scholar]
- Rees MI, Andrew M, Jawad S, Owen MJ. Evidence for recessive as well as dominant forms of startle disease (hyperekplexia) caused by mutations in the alpha 1 subunit of the inhibitory glycine receptor. Hum Mol Genet. 1994;3:2175–2179. doi: 10.1093/hmg/3.12.2175. [DOI] [PubMed] [Google Scholar]
- Rees MI, Lewis TM, Kwok JB, Mortier GR, Govaert P, Snell RG, Schofield PR, Owen MJ. Hyperekplexia associated with compound heterozygote mutations in the beta-subunit of the human inhibitory glycine receptor (GLRB) Hum Mol Genet. 2002;11:853–860. doi: 10.1093/hmg/11.7.853. [DOI] [PubMed] [Google Scholar]
- Rees MI, Harvey K, Pearce BR, Chung SK, Duguid IC, Thomas P, Beatty S, Graham GE, Armstrong L, Shiang R, Abbott KJ, Zuberi SM, Stephenson JB, Owen MJ, Tijssen MA, van den Maagdenberg AM, Smart TG, Supplisson S, Harvey RJ. Mutations in the gene encoding GlyT2 (SLC6A5) define a presynaptic component of human startle disease. Nat Genet. 2006;38:801–806. doi: 10.1038/ng1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SG, Buckwalter MS, Lynch JW, Handford CA, Segura L, Shiang R, Wasmuth JJ, Camper SA, Schofield P, O'Connell P. A missense mutation in the gene encoding the alpha 1 subunit of the inhibitory glycine receptor in the spasmodic mouse. Nat Genet. 1994;7:131–135. doi: 10.1038/ng0694-131. [DOI] [PubMed] [Google Scholar]
- Sadtler S, Laube B, Lashub A, Nicke A, Betz H, Schmalzing G. A basic cluster determines topology of the cytoplasmic M3–M4 loop of the glycine receptor alpha1 subunit. J Biol Chem. 2003;278:16782–16790. doi: 10.1074/jbc.M213077200. [DOI] [PubMed] [Google Scholar]
- Saul B, Schmieden V, Kling C, Mülhardt C, Gass P, Kuhse J, Becker CM. Point mutation of glycine receptor alpha 1 subunit in the spasmodic mouse affects agonist responses. FEBS Lett. 1994;350:71–76. doi: 10.1016/0014-5793(94)00736-5. [DOI] [PubMed] [Google Scholar]
- Schorge S, Colquhoun D. Studies of NMDA receptor function and stoichiometry with truncated and tandem subunits. J Neurosci. 2003;23:1151–1158. doi: 10.1523/JNEUROSCI.23-04-01151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüler T, Mesic I, Madry C, Bartholomäus I, Laube B. Formation of NR1/NR2 and NR1/NR3 heterodimers constitutes the initial step in N-methyl-d-aspartate receptor assembly. J Biol Chem. 2008;283:37–46. doi: 10.1074/jbc.M703539200. [DOI] [PubMed] [Google Scholar]
- Sontheimer H, Becker CM, Pritchett DB, Schofield PR, Grenningloh G, Kettenmann H, Betz H, Seeburg PH. Functional chloride channels by mammalian cell expression of rat glycine receptor subunit. Neuron. 1989;2:1491–1497. doi: 10.1016/0896-6273(89)90195-5. [DOI] [PubMed] [Google Scholar]
- van Geest M, Lolkema JS. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol Mol Biol Rev. 2000;64:13–33. doi: 10.1128/mmbr.64.1.13-33.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villmann C, Bull L, Hollmann M. Kainate binding proteins possess functional ion channel domains. J Neurosci. 1997;17:7634–7643. doi: 10.1523/JNEUROSCI.17-20-07634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villmann C, Hoffmann J, Werner M, Kott S, Strutz-Seebohm N, Nilsson T, Hollmann M. Different structural requirements for functional ion pore transplantation suggest different gating mechanisms of NMDA and kainate receptors. J Neurochem. 2008;107:453–465. doi: 10.1111/j.1471-4159.2008.05623.x. [DOI] [PubMed] [Google Scholar]
- Wallace DJ, Zum Alten Borgloh SM, Astori S, Yang Y, Bausen M, Kugler S, Palmer AE, Tsien RY, Sprengel R, Kerr JN, Denk W, Hasan MT. Single-spike detection in vitro and in vivo with a genetic Ca2+ sensor. Nat Methods. 2008;5:797–804. doi: 10.1038/nmeth.1242. [DOI] [PubMed] [Google Scholar]
- Yevenes GE, Moraga-Cid G, Guzmán L, Haeger S, Oliveira L, Olate J, Schmalzing G, Aguayo LG. Molecular determinants for G protein βγ modulation of ionotropic glycine receptors. J Biol Chem. 2006;281:39300–39307. doi: 10.1074/jbc.M608272200. [DOI] [PubMed] [Google Scholar]
- Young TL, Cepko CL. A role for ligand-gated ion channels in rod photoreceptor development. Neuron. 2004;41:867–879. doi: 10.1016/s0896-6273(04)00141-2. [DOI] [PubMed] [Google Scholar]
- Zolotukhin S, Potter M, Zolotukhin I, Sakai Y, Loiler S, Fraites TJ, Jr, Chiodo VA, Phillipsberg T, Muzyczka N, Hauswirth WW, Flotte TR, Byrne BJ, Snyder RO. Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods. 2002;28:158–167. doi: 10.1016/s1046-2023(02)00220-7. [DOI] [PubMed] [Google Scholar]